Abstract
While lipid droplets have traditionally been considered as inert sites for the storage of triacylglycerols and sterol esters, they are now recognized as dynamic and functionally diverse organelles involved in energy homeostasis, lipid signaling, and stress responses. Unlike most other organelles, lipid droplets are delineated by a half-unit membrane whose protein constituents are poorly understood, except in the specialized case of oleosins, which are associated with seed lipid droplets. Recently, we identified a new class of lipid-droplet associated proteins called LDAPs that localize specifically to the lipid droplet surface within plant cells and share extensive sequence similarity with the small rubber particle proteins (SRPPs) found in rubber-accumulating plants. Here, we provide additional evidence for a role of LDAPs in lipid accumulation in oil-rich fruit tissues, and further explore the functional relationships between LDAPs and SRPPs. In addition, we propose that the larger LDAP/SRPP protein family plays important roles in the compartmentalization of lipophilic compounds, including triacylglycerols and polyisoprenoids, into lipid droplets within plant cells. Potential roles in lipid droplet biogenesis and function of these proteins also are discussed.
Lipid droplets play an essential role in the life cycle of plants by housing lipid storage compounds, usually triacylglycerols, in seeds that are mobilized to support post-germinative growth, prior to photosynthetic establishment. Given this critical role in plant growth and development, the majority of research on plant lipid droplets has focused on their function in seed tissues. For instance, purification of lipid droplets from plant oilseeds resulted in the identification and characterization of the oleosins, which are an abundant class of lipid droplet-surface-associated proteins important for stabilizing lipid droplets during seed desiccationCitation1-Citation3 and possibly serving as sites for recruitment of lipases that facilitate the breakdown of stored triacylglycerols during seedling establishment.Citation4
It is now appreciated, however, that lipid droplets have numerous functions beyond lipid storage in seeds and that they are present in nearly all plant cell types, many of which do not accumulate appreciable amounts of lipid, such as the cells in leaves, stems, and roots.Citation5 There is also emerging evidence that lipid droplets are highly dynamic organelles involved in a variety of cellular processes and physiological responses, some of which appear to be conserved among eukaryotes.Citation6-Citation8 Nevertheless, the precise functions of lipid droplets in non-seed cell types in plants are currently poorly understood.
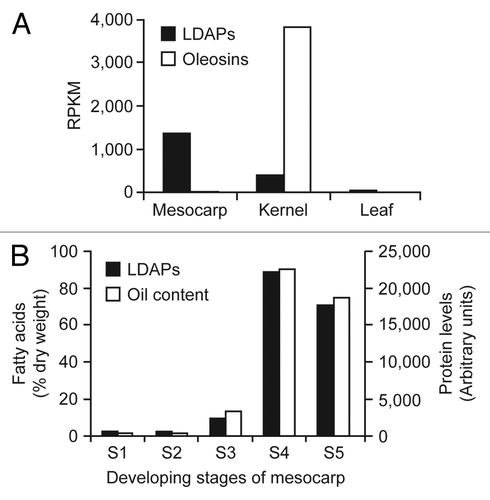
In an effort to increase our understanding of lipid droplet biogenesis and functions in plants, we recently characterized the proteome of lipid droplets isolated from the mesocarp of avocado (Persea americana).Citation9 This tissue was selected for analysis since it is a rich source of non-seed lipid droplets that lack the abundant oleosins found in oilseed tissues. Briefly, proteins enriched in the isolated avocado mesocarp lipid droplet fraction were identified using a combination of multi-dimensional protein identification technology and peptide mass fingerprinting, using an avocado RNAseq-derived “proteome” for query. Two of the most abundant proteins associated with these lipid droplets were highly similar (86%) in sequence to each other and, thus, were annotated as lipid droplet-associated protein 1 (LDAP1) and LDAP2 (; Pam_LDAP1 and Pam_LDAP2). We also showed previously that LDAP1 and LDAP2 gene expression during development of avocado mesocarp increased in correlation with oil accumulation.Citation9 Interestingly, transcriptome analysis of various tissues of oil palm (Elaeis guineensis) revealed the presence of three LDAP-like genes, of which one of them showed the highest homology with LDAPs from other species (; Egu_LDAP-like). The transcript levels for these LDAPs are high in oil palm mesocarp, another oil-rich non-seed tissue, during the period of oil accumulation ().Citation10 The expression of the LDAP genes is moderate in oil palm kernel, which accumulates about 40–60% oil, but was not elevated in leaf tissues that do not accumulate high amounts of oil (). It is worth pointing out that transcript levels for oleosins were also abundant in oil palm kernel but absent in mesocarp, where LDAP expression is more predominant (). Most strikingly, proteome analysis shows that LDAP levels match closely to that of oil accumulation during mesocarp development (). Taken together, these observations support a role for LDAP-like proteins in the biogenesis of lipid droplets and accumulation of triacylglycerol in lipid-rich tissues such as avocado and oil palm mesocarp, where oleosin-mediated stabilization of lipid droplets is absent.
Figure 1. Sequence analysis of LDAP-like proteins. (A) ClustalW alignment of bona fide lipid droplet-associated proteins (LDAPs) from avocado (Pam_LDAP1 and Pam_LDAP2) and Arabidopsis (Ath_LDAP), and biochemically characterized small rubber particle proteins (SRPPs) from rubber tree (Hbr_SRPP) and guayule (Par_SRPP). GenBank accession numbers are KF031141, KF031142, NM_111423, AJ223388, and AAQ11374, respectively. The LDAP-like sequence (M01000058007; available at http://www.biomemb.cnrs.fr/contigs.html) from oil palm (E. guineensis), Egu_LDAP-like was identified by TBLASTN search of its transcriptome dataCitation10 using Ath_LDAP as a query. (B) Cartoon illustrating the various forms and functional domains fused to LDAPs in certain plant species. (1) LDAP domain, which is shared among LDAPs and SRPPs in non-rubber-accumulating and rubber-accumulating plants, respectively (see A for examples). (2) Rubber elongation factor (REF) proteins, which are similar in sequence to the N-terminal portion of LDAPs and SRPPs and have been shown to associate with rubber particles isolated from the rubber tree (Hevea brasiliensis; e.g., GenBank number X56535).Citation14 (3) Two similar genes, at 2 different loci in apple (Malus domestica; Phytozome loci MDP0000557646 and MDP0000608906 [www.phytozome.net]), encoding putative proteins that each have a RALF (Rapid Alkalinization Factor) domain fused at the N terminus of the LDAP domain. RALF domains are peptide hormones involved in various aspects of plant growth and development.Citation16 (4) C-terminal domain (CTD) small phosphatase-like protein 2 sequences fused to both N- and C-terminal sides of an LDAP in Medicago truncatula (Phytozome locus Medtr3g085400). (5) HORMA domain (named after the Hop1p, Rev7p, and MAD2 proteins), typically involved in protein–protein interactions associated with chromatin binding,Citation17 fused to LDAP in flax (Linum usitatissimum; Phytozome locus Lus10015786.g). (6) CTD small phosphatase-like protein 2 sequence and a HORMA domain fused to LDAP in both flax (Phytozome locus Lus10037019.g) and strawberry (Fragaria vesca; GenBank number XP_004310215). (7) Three LDAP domains fused together in a single gene in cotton (Gossypium raimondii; Phytozome locus Gorai.007G341500). Notably, we could amplify cDNA fragments corresponding to this latter gene, including one that spanned portions of all 3 LDAP domains (data not shown), confirming it is an authentic fusion.
![Figure 1. Sequence analysis of LDAP-like proteins. (A) ClustalW alignment of bona fide lipid droplet-associated proteins (LDAPs) from avocado (Pam_LDAP1 and Pam_LDAP2) and Arabidopsis (Ath_LDAP), and biochemically characterized small rubber particle proteins (SRPPs) from rubber tree (Hbr_SRPP) and guayule (Par_SRPP). GenBank accession numbers are KF031141, KF031142, NM_111423, AJ223388, and AAQ11374, respectively. The LDAP-like sequence (M01000058007; available at http://www.biomemb.cnrs.fr/contigs.html) from oil palm (E. guineensis), Egu_LDAP-like was identified by TBLASTN search of its transcriptome dataCitation10 using Ath_LDAP as a query. (B) Cartoon illustrating the various forms and functional domains fused to LDAPs in certain plant species. (1) LDAP domain, which is shared among LDAPs and SRPPs in non-rubber-accumulating and rubber-accumulating plants, respectively (see A for examples). (2) Rubber elongation factor (REF) proteins, which are similar in sequence to the N-terminal portion of LDAPs and SRPPs and have been shown to associate with rubber particles isolated from the rubber tree (Hevea brasiliensis; e.g., GenBank number X56535).Citation14 (3) Two similar genes, at 2 different loci in apple (Malus domestica; Phytozome loci MDP0000557646 and MDP0000608906 [www.phytozome.net]), encoding putative proteins that each have a RALF (Rapid Alkalinization Factor) domain fused at the N terminus of the LDAP domain. RALF domains are peptide hormones involved in various aspects of plant growth and development.Citation16 (4) C-terminal domain (CTD) small phosphatase-like protein 2 sequences fused to both N- and C-terminal sides of an LDAP in Medicago truncatula (Phytozome locus Medtr3g085400). (5) HORMA domain (named after the Hop1p, Rev7p, and MAD2 proteins), typically involved in protein–protein interactions associated with chromatin binding,Citation17 fused to LDAP in flax (Linum usitatissimum; Phytozome locus Lus10015786.g). (6) CTD small phosphatase-like protein 2 sequence and a HORMA domain fused to LDAP in both flax (Phytozome locus Lus10037019.g) and strawberry (Fragaria vesca; GenBank number XP_004310215). (7) Three LDAP domains fused together in a single gene in cotton (Gossypium raimondii; Phytozome locus Gorai.007G341500). Notably, we could amplify cDNA fragments corresponding to this latter gene, including one that spanned portions of all 3 LDAP domains (data not shown), confirming it is an authentic fusion.](/cms/asset/04651e21-47fd-4c20-842d-fa772e9d8e37/kpsb_a_10927141_f0001.gif)
Figure 2. Expression pattern of LDAPs in oil palm (E. guineensis). (A) Three LDAP-like genes, represented by 5 contigs (M01000063962, M01000058007, M01000030686, M01000019267, and M01000000234), were identified in oil palm. The average transcript levels of the 3 LDAPs were higher in mesocarp than kernel, which was harvested 15 wk after pollination, during which time oil accumulation is at a maximum. On the contrary, average EST levels for oleosins (sum of GBAF147758 and GAJH01049154) were highest in kernel and very low in mesocarp. Both LDAP and oleosin genes were barely expressed in leaf tissues. Transcriptome data, in raw format, are available in GenBank Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) (oil palm: SRX059258–62). (B) Temporal changes in protein accumulation for the 3 LDAPs during mesocarp development of oil palm, showing a rapid increase in abundance during the period when oil accumulation occurs (stages S3 to S5). The developmental stages of the mesocarp (S1-S5) were previously described.Citation10 Relative protein levels from oil palm mesocarp were estimated by label-free proteomics (Dupuy JW and Arondel V, unpublished).
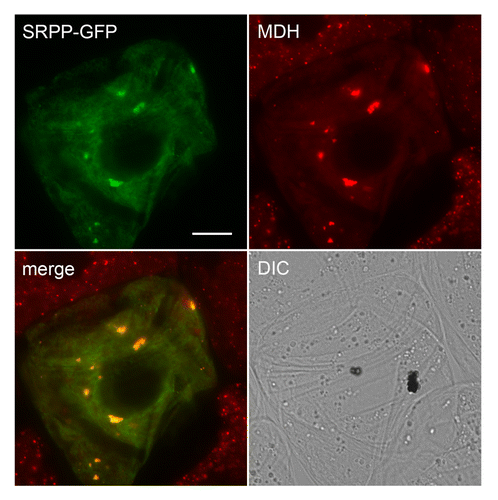
A search for LDAP-like genes in eukaryotic organisms whose genomes have been fully sequenced revealed that this gene is plant-specific and is highly conserved among all plant species, and with most plants having 3 different LDAP-like genes.Citation9 The LDAP proteins are also highly similar in sequence to small rubber particle proteins (SRPPs) that accumulate in certain rubber-producing plants, such as rubber tree (Hevea brasiliensis)Citation11 and guayule (Parthenium argentatum)Citation12 (; Hbr_SRPP and Par_SRPP, respectively). Thus, it appears that the LDAP genes are not unique to plants that produce high amounts of oil in their mesocarp, but rather are more likely involved in conserved aspects of lipid droplet biogenesis that are shared among most plant species. In support of this premise, we showed previously that the protein from Arabidopsis thaliana (At3g05500) with highest homology to the avocado LDAPs (; Ath_LDAP) did indeed target specifically to lipid droplets in model, non-seed plant cells, i.e., tobacco suspension-cultured cells.Citation9 At3g05500 is also highly expressed in developing Arabidopsis seeds with a temporal pattern similar to oil body biogenesis and oleosin accumulation (Toronto BAR eFP browser; http://bar.utoronto.ca/), consistent with a role in both seeds and non-seed tissues. Here, we show that guayule SRPP, which is associated with lipid droplets containing polyisoprenoids,Citation12 is also capable of targeting to triacylglycerol-containing lipid droplets in tobacco cells (). Given the similar targeting of LDAPs and SRPPs to lipid droplets containing triacylglycerol, it is possible that the LDAP/SRPP family of proteins share a generalized role in lipid droplet biogenesis by binding to and stabilizing the lipid droplet surface, thereby promoting the proper partitioning of the lipophilic compounds stored within. Evidence in support of this idea is that knock down of SRPP gene expression in Russian dandelion (Taraxacum brevicorniculatum) results in rubber particles that are less stable and tend to aggregate, resulting in an overall reduction in rubber production.Citation13 Whether the knock down of LDAPs in a non-rubber accumulating plant, such as Arabidopsis, yields a similar phenotype, however, remains to be determined.
Figure 3. Localization of guayule SRPP-GFP to lipid droplets in a tobacco cell. Shown are representative epifluorescence micrographs of tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) suspension-cultured cells, which serve as a well-characterized system for studying protein localization in plant cells.Citation18 BY-2 cells were transiently transformed via biolistic bombardment with plasmid DNA encoding full-length guayule SRPPCitation12 (GenBank number AF541942) C-terminally fused to the N terminus of the green fluorescent protein (SRPP-GFP). Following bombardment, cells were incubated in linoleic acid, which induces a dramatic increase in the number and size of lipid droplets in these cells (see ref. Citation9 for additional details), and then incubated with monodansylpentane (MDH), which is a blue-fluorescing marker dye for lipid droplets in living cells.Citation19 Note that the fluorescence attributable to the MDH-stained lipid droplets is false colorized red. The yellow color in the merged images represents obvious co-localizations between SRPP-GFP and MDH-stained lipid droplets, most of which have coalesced, apparently due to the ectopic (over)expression of the fusion protein. These larger coalesced structures are not observed in the neighboring non-transformed cells wherein lipid droplets are usually dispersed throughout the cytosol. Similar coalescence of lipid droplets was observed in BY-2 cells transiently overexpressing Arabidopsis LDAP (Gidda SK, Watt SC, and Mullen RT, unpublished), as well as in various other cells types in which other lipid droplet proteins, such as perilipin 1 and the ancient ubiquitous protein 1, are ectopically (over)expressed.Citation20,Citation21 Shown also is the corresponding differential interference contrast (DIC) image. Bar = 10 μm.
Although the precise functions of LDAPs are not yet fully understood, the SRPPs are known to function by stimulating the synthesis of polyisoprenoids in isolated rubber particles.Citation11 Interestingly, rubber particles isolated from H. brasiliensis also contain shorter SRPP-like proteins called the rubber elongation factors (REFs), which also stimulate rubber production.Citation14,Citation15 The REF proteins are highly similar to the N-terminal regions of SRPPs and LDAPs (), and thus may represent a minimal lipid droplet-associating domain. It is also notable that while all higher plants are known to have the longer LDAP- or SRPP-like genes, only a few plants whose genomes have been sequenced contain predicted REF-like genes, including grape, rice, maize, and eucalyptus (data not shown). In addition to these shorter REF-like genes, certain plant species contain significantly longer LDAP-like genes that encode a fusion of LDAP to other domains, such as phosphatases, HORMA-like domains, or RALF-33-like peptide hormones, and also a fusion consisting of 3 LDAPs joined in tandem (). However, whether any of these represent bona fide functional genes or artifacts of genome annotation requires further investigation. Nonetheless, it will be interesting to further elucidate the role(s) of the LDAPs in lipid droplet ontogeny and regulation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by a grant from the US Department of Energy, BER Division, DE-FG02–09ER64812/DE-SC0000797 and by the US Department of Energy, Great Lakes Bioenergy Research Center, Cooperative Agreement DE-FC02–07ER6449. The authors thank Grisel Ponciano (USDA-ARS Western Regional Research Center, Albany, CA) for providing the guayule SRRP-GFP plasmid. Proteomic data were analyzed by JW Dupuy at the Proteome Platform of Functional Genomic Center of Bordeaux, France.
References
- Huang AHC. Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol 1992; 43:177 - 200; http://dx.doi.org/10.1146/annurev.pp.43.060192.001141
- Frandsen GI, Mundy J, Tzen JT. Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 2001; 112:301 - 7; http://dx.doi.org/10.1034/j.1399-3054.2001.1120301.x; PMID: 11473685
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 2006; 18:1961 - 74; http://dx.doi.org/10.1105/tpc.106.041269; PMID: 16877495
- Quettier AL, Eastmond PJ. Storage oil hydrolysis during early seedling growth. Plant Physiol Biochem 2009; 47:485 - 90; http://dx.doi.org/10.1016/j.plaphy.2008.12.005; PMID: 19136267
- Chapman KD, Dyer JM, Mullen RT. Biogenesis and functions of lipid droplets in plants: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man. J Lipid Res 2012; 53:215 - 26; http://dx.doi.org/10.1194/jlr.R021436; PMID: 22045929
- van der Schoot C, Paul LK, Paul SB, Rinne PL. Plant lipid bodies and cell-cell signaling: a new role for an old organelle?. Plant Signal Behav 2011; 6:1732 - 8; http://dx.doi.org/10.4161/psb.6.11.17639; PMID: 22057325
- Murphy DJ. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 2012; 249:541 - 85; http://dx.doi.org/10.1007/s00709-011-0329-7; PMID: 22002710
- Walther TC, Farese RV Jr.. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 2012; 81:687 - 714; http://dx.doi.org/10.1146/annurev-biochem-061009-102430; PMID: 22524315
- Horn PJ, James CN, Gidda SK, Kilaru A, Dyer JM, Mullen RT, Ohlrogge JB, Chapman KD. Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol 2013; 162:1926 - 36; http://dx.doi.org/10.1104/pp.113.222455; PMID: 23821652
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue GF, Drira N, Ohlrogge JB, Arondel V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci U S A 2011; 108:12527 - 32; http://dx.doi.org/10.1073/pnas.1106502108; PMID: 21709233
- Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KH. Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis.. J Biol Chem 1999; 274:17132 - 8; http://dx.doi.org/10.1074/jbc.274.24.17132; PMID: 10358068
- Kim IJ, Ryu SB, Kwak YS, Kang H. A novel cDNA from Parthenium argentatum Gray enhances the rubber biosynthetic activity in vitro. J Exp Bot 2004; 55:377 - 85; http://dx.doi.org/10.1093/jxb/erh039; PMID: 14718497
- Hillebrand A, Post JJ, Wurbs D, Wahler D, Lenders M, Krzyzanek V, Prüfer D, Gronover CS. Down-regulation of small rubber particle protein expression affects integrity of rubber particles and rubber content in Taraxacum brevicorniculatum.. PLoS One 2012; 7:e41874; http://dx.doi.org/10.1371/journal.pone.0041874; PMID: 22911861
- Dennis MS, Henzel WJ, Bell J, Kohr W, Light DR. Amino acid sequence of rubber elongation factor protein associated with rubber particles in Hevea latex. J Biol Chem 1989; 264:18618 - 26; PMID: 2808390
- Dennis MS, Light DR. Rubber elongation factor from Hevea brasiliensis. Identification, characterization, and role in rubber biosynthesis. J Biol Chem 1989; 264:18608 - 17; PMID: 2681199
- Cao J, Shi F. Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol Bioinform Online 2012; 8:271 - 92; http://dx.doi.org/10.4137/EBO.S9652; PMID: 22745530
- Ferdous M, Higgins JD, Osman K, Lambing C, Roitinger E, Mechtler K, Armstrong SJ, Perry R, Pradillo M, Cuñado N, et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet 2012; 8:e1002507; http://dx.doi.org/10.1371/journal.pgen.1002507; PMID: 22319460
- Brandizzi F, Irons S, Kearns A, Hawes C. BY-2 cells: culture and transformation for live cell imaging. Curr Protoc Cell Biol 2003; Chapter 1:7; http://dx.doi.org/10.1002/0471143030.cb0107s19; PMID: 18228413
- Yang H-J, Hsu CL, Yang J-Y, Yang WY. Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS One 2012; 7:e32693; http://dx.doi.org/10.1371/journal.pone.0032693; PMID: 22396789
- Lohmann D, Spandl J, Stevanovic A, Schoene M, Philippou-Massier J, Thiele C. Monoubiquitination of ancient ubiquitous protein 1 promotes lipid droplet clustering. PLoS One 2013; 8:e72453; http://dx.doi.org/10.1371/journal.pone.0072453; PMID: 24039768
- Orlicky DJ, Monks J, Stefanski AL, McManaman JL. Dynamics and molecular determinants of cytoplasmic lipid droplet clustering and dispersion. PLoS One 2013; 8:e66837; http://dx.doi.org/10.1371/journal.pone.0066837; PMID: 23825572