Abstract
Low-fluence and long-wavelength UV-B light promotes photomorphogenic development in Arabidopsis. CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) is a positive regulator in this pathway while it is a negative regulator of the traditional photomorphogenesis triggered by far-red and visible light. We have recently reported the mechanism by which the switch of COP1 function is accomplished in distinct light contexts. In response to photomorphogenic UV-B, the photoactivated UV RESISTANCE LOCUS 8 (UVR8) associates with the COP1- SUPRESSOR OF PHYA (SPA) core complexes, resulting in the physical and functional disassociation of COP1-SPA from the CULLIN4-DAMAGED DNA BINDING PROTEIN 1 (CUL4-DDB1) E3 scaffold. These UV-B dependent UVR8-COP1-SPA complexes promote the stability and activity of ELONGATED HYPOCOTYL 5 (HY5), and eventually cause COP1 to switch from repressing to promoting photomorphogenesis. In addition, it is possible that CUL4-DDB1 might simultaneously recruit alternative DDB1 BINDING WD40 (DWD) proteins to repress this UV-B-specific signaling. Further investigation is required, however, to verify this hypothesis. Overall, the coordinated organization of various protein complexes facilitates an efficient and balanced UV-B signaling.
Keywords: :
To cope with the changing light environment, higher plants have evolved the capacity to sense and interpret diverse light signals, in order to optimize their growth. UV-B light (280—315 nm) comprises a minor proportion of sunlight that reaches the earth surface, and is processed by plants not only as a damaging stimulus, but also as an informational signal. Specifically, high-fluence and short-wavelength UV-B light induces stress-related physiological responses, while low-fluence and long-wavelength UV-B light promotes photomorphogenic development.Citation1
UV RESISTANCE LOCUS 8 (UVR8) is a UV-B light receptor that has been recently identified in Arabidopsis thaliana. Upon irradiation with photomorphogenic UV-B, the dark-state dimeric UVR8 perceives UV-B light via its own tryptophan residues, which acts as internal chromophores, and monomerizes by disrupting the critical intermolecular bonds shaped by its own arginine residues.Citation2,Citation3 The lit-state monomeric UVR8 then rapidly interacts with CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) in order to trigger downstream signaling.Citation4,Citation5
COP1 is an evolutionally conserved and multifunctional RING E3 ubiquitin ligase. It was originally identified by screening Arabidopsis seedlings that exhibit constitutive photomorphogenesis in darkness.Citation6 Molecular and biochemical studies have revealed that COP1 functions in COP1-SUPRESSOR OF PHYA (SPA) complexes.Citation7 They serve as substrate receptors for the CULLIN4- DAMAGED DNA BINDING PROTEIN 1 (CUL4-DDB1) E3 apparatus, to target photomorphogenesis promoting transcription factors including ELONGATED HYPOCOTYL 5 (HY5) for the ubiquitin-proteasome system (UPS)-mediated degradation.Citation8 Thus, COP1 is a central repressor of the traditional photomorphogenesis triggered by far-red and visible light.Citation9
Intriguingly, COP1 has also been shown to be a positive regulator of UV-B-induced photomorphogenesis,Citation4,Citation10 and to no longer act antagonistically against HY5 under UV-B.Citation11,Citation12 The loss-of-function mutation of either COP1 or HY5 suffers from abolished transcriptomic responses to photomorphogenic UV-B, and consequently results in defective UV-B-induced photomorphogenesis and UV-B acclimation.Citation10 In addition to these discoveries, several additional observations listed below prompted us to investigate the mechanism of COP1’s role in this UV-B specific signaling, particularly with regard to the changes induced in the relevance interaction between COP1 and HY5. Both COP1 and HY5 show nuclear localization under UV-B.Citation10 COP1 is required for the UV-B-mediated HY5 induction,Citation4 and HY5 activates COP1 in a positive feedback by targeting the COP1 promoter.Citation12
We found that under photomorphogenic UV-B, monomerized UVR8 sequestered COP1 from DDB1, which in turn caused COP1-SPA to disassociate from CUL4-DDB1. In agreement with this physical segregation, COP1-SPA also displayed functional disassociation from CUL4-DDB1, and CUL4 was identified as a repressor of UV-B-induced photomorphogenesis.Citation11
In contrast, we showed that COP1 and SPA proteins remained tightly associated with one another regardless of the presence or absence of UV-B. Under photomorphogenic UV-B, UVR8 was observed to associate with the COP1-SPA core to constitute novel complexes. We then verified that SPA proteins, like COP1 and UVR8, played positive regulatory roles in UV-B-induced photomorphogenesis. More importantly, we demonstrated that the UV-B-dependent UVR8-COP1-SPA complexes promoted the stability and activity of HY5, which enabled a functional switch of COP1 from repressing to promoting photomorphogenesis. In YFP-UVR8W285A transgenic plants, the constitutive UVR8-COP1-SPA complexes were found to continuously facilitate HY5 stability and activity, and to eventually drive the Arabidopsis seedlings to develop constitutive photomorphogenesis in darkness.Citation11 However, how HY5 is stabilized is a subject that awaits further investigation. Our results have indicated that an alternative E3 ubiquitin ligase might be responsible for the turnover of HY5 protein, and UVR8-COP1-SPA complexes might deactivate this E3 ligase. Also, it has been pointed out that the stability and activity of HY5 is modulated by its phosphorylation.Citation13 Thus a close examination of the phosphorylation or/and other modifications of HY5 might reveal that how HY5 is regulated post-transcriptionally by photomorphogenic UV-B.
As it does under other light conditions, CUL4 represses photomorphogenesis under UV-B.Citation11,Citation14 Nevertheless, it remains unknown, regarding the mechanism by which CUL4 plays a repressive role in UV-B-induced photomorphogenesis. It has been extensively studied that DDB1 is an adaptor protein that recruits a group of proteins named DDB1-CUL4 ASSOCIATED FACTOR (DCAF) as substrate receptors for CUL4-based E3 ligase in both animals and plants.Citation15-Citation19 In Arabidopsis, DCAFs, also designated as DWD proteins, are functionally involved in a wide variety of developmental processes.Citation19 Therefore, it is reasonable that one or more unknown DWD proteins might join the CUL4-DDB1 E3 scaffold to repress UV-B-induced photomorphogenesis.
Two previously identified negative regulators in this pathway, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2,Citation20 are potential DWD proteins as each of them possesses one typical WDXR motif that might mediate their direct interaction with DDB1. Furthermore, multiple lines of evidence indicate that CUL4-DDB1 and RUP1/RUP2 might function in concert. For example, a reduction in CUL4 or RUP1/RUP2 function was observed to result in hypersensitivity to photomorphogenic UV-B in Arabidopsis seedlings, which were found to display enhanced induction of UV-B-responsive genes and increased accumulation of HY5 protein, the known UV-B signaling hub.Citation11,Citation20 Thus, further analyses of the genetic and biochemical interaction between CUL4 and RUP1/RUP2 might assist substantiating this hypothesis. Since RUP1 and RUP2 are known to facilitate the conformational reversion of UVR8,Citation21,Citation22 it is also of interest and significance to investigate whether or not CUL4 participates in this process.
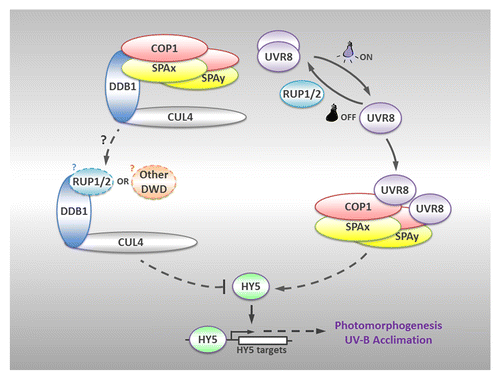
Taken together, we propose a model for the organization of protein complexes under photomorphogenic UV-B (). Following UVR8-mediated UV-B light perception, monomerized UVR8 directly interacts with COP1 so as to form UVR8-COP1-SPA complexes, which, in turn, promote the stability and activity of HY5. HY5 is then critically involved in downstream transcriptional regulation. On the other hand, when sequestered by UVR8 monomers, COP1-SPA core complexes disassociate from the CUL4-DDB1 E3 apparatus which might then recruit alternative DWD proteins to repress UV-B-induced photomorphogenesis. Overall, these complexes are structurally and functionally coordinated to ensure an efficient and balanced photomorphogenic UV-B signaling.
Figure 1. A model for the organization of protein complexes under photomorphogenic UV-B. Upon UV-B irradiation, UVR8 switches from dimer to monomer, and directly interacts with COP1 to form UVR8-COP1-SPA complexes, which promote the stability and activity of HY5. HY5 positively regulates downstream transcriptional responses so as to facilitate UV-B-induced photomorphogenesis and acclimation. On the other hand, the CUL4-DDB1 E3 apparatus, which releases the COP1-SPA core complexes sequestered by UVR8 monomers, might recruit alternative DWD proteins such as RUP1/RUP2 to repress UV-B-induced photomorphogenesis. Once the UV-B irradiation is removed, RUP1 and RUP2 mediate the redimerization of UVR8. Solid lines indicate direct effects, while dotted lines represent indirect regulation. Question marks denote the hypothesis, which awaits further investigation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Abigail Coplin for her critical reading and commenting on the manuscript.
References
- Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 2009; 60:407 - 31; http://dx.doi.org/10.1146/annurev.arplant.59.032607.092953; PMID: 19400728
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 2012; 335:1492 - 6; http://dx.doi.org/10.1126/science.1218091; PMID: 22323738
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al. Structural basis of ultraviolet-B perception by UVR8. Nature 2012; 484:214 - 9; http://dx.doi.org/10.1038/nature10931; PMID: 22388820
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 2009; 28:591 - 601; http://dx.doi.org/10.1038/emboj.2009.4; PMID: 19165148
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011; 332:103 - 6; http://dx.doi.org/10.1126/science.1200660; PMID: 21454788
- Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 2005; 15:618 - 25; http://dx.doi.org/10.1016/j.tcb.2005.09.007; PMID: 16198569
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 2008; 20:2307 - 23; http://dx.doi.org/10.1105/tpc.107.056580; PMID: 18812498
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 2010; 22:108 - 23; http://dx.doi.org/10.1105/tpc.109.065490; PMID: 20061554
- Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 2012; 17:584 - 93; http://dx.doi.org/10.1016/j.tplants.2012.05.004; PMID: 22705257
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 2006; 18:1975 - 90; http://dx.doi.org/10.1105/tpc.105.040097; PMID: 16829591
- Huang X, Ouyang X, Yang P, Lau OS, Chen L, Wei N, Deng XW. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci U S A 2013; 110:16669 - 74; http://dx.doi.org/10.1073/pnas.1316622110; PMID: 24067658
- Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 2012; 24:4590 - 606; http://dx.doi.org/10.1105/tpc.112.103994; PMID: 23150635
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 2000; 19:4997 - 5006; http://dx.doi.org/10.1093/emboj/19.18.4997; PMID: 10990463
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell 2006; 18:1991 - 2004; http://dx.doi.org/10.1105/tpc.106.043224; PMID: 16844902
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev 2006; 20:2949 - 54; http://dx.doi.org/10.1101/gad.1483206; PMID: 17079684
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006; 443:590 - 3; PMID: 16964240
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709 - 21; http://dx.doi.org/10.1016/j.molcel.2006.08.010; PMID: 16949367
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 2006; 5:1675 - 80; http://dx.doi.org/10.4161/cc.5.15.3149; PMID: 16861906
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 2008; 20:152 - 67; http://dx.doi.org/10.1105/tpc.107.055418; PMID: 18223036
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci U S A 2010; 107:20132 - 7; http://dx.doi.org/10.1073/pnas.0914532107; PMID: 21041653
- Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci U S A 2013; 110:1113 - 8; http://dx.doi.org/10.1073/pnas.1214237110; PMID: 23277547
- Heilmann M, Jenkins GI. Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol 2013; 161:547 - 55; http://dx.doi.org/10.1104/pp.112.206805; PMID: 23129206