Abstract
Posttranslational modification of proteins by SUMO plays essential roles in plant growth and development. We, and others, have previously identified Arabidopsis proteins covalently modified by SUMO. In our recent report, we assessed the extent and significance of non-covalent SUMO interactions with plant proteins by using three Arabidopsis SUMO isoforms as baits in large-scale yeast two-hybrid screens. We identified six proteins that regulate the reversible methylation and demethylation of histones and DNA, and six proteins that we showed to be the plant homologs of SUMO-Targeted Ubiquitin E3 Ligases (STUbLs). This implicates SUMO in a variety of developmental programs including floral transition, genome imprinting, and transcriptional control of a large number of genes. Intriguingly, whereas only two STUbLs were identified in other organisms, the identification of six STUbLs in Arabidopsis is consistent with the more complex repertoire of genes regulating the SUMO system in plants. Some Arabidopsis STUbLs appear to have retained roles conserved throughout eukaryotes, whereas others may have evolved novel plant functions. AT-STUbL4, for example, contributes to the floral transition by reducing the levels of the floral repressor Cycling Dof Factor 2 (CDF2). I discuss our findings and the potential they provide to study the role of SUMO in plant development.
Ubiquitin (Ub), and the Small Ub-like MOdifer (SUMO) are likely the two most important Ub-fold proteins. Whereas Ub functions primarily as a “death” signal that targets proteins for degradation by the 26S proteasome, no direct evidence has implicated SUMO in protein degradation.Citation1 Rather, SUMO modification of proteins modulates protein structure and function in a variety of ways that impact cellular and organismal development. In Arabidopsis, SUMO1 and SUMO2 double knockout mutants are embryo-lethal, and impairing the basic SUMO conjugation machinery (the SUMO Activating Enzyme, SAE or E1; or the SUMO Conjugating Enzyme, SCE or E2) also leads to embryonic lethality, suggesting that protein SUMOylation is essential for plant growth and development.Citation2 Mutations of the genes encoding SUMO proteases (e.g., ESD4) or SUMO E3 ligases (e.g., SIZ1 or HPY2/MMS21) cause major developmental problems as well.Citation3-Citation5 These proteins regulate the dynamic levels of SUMOylated proteins in the cell, and hence maintaining proper levels of SUMOylation is also crucial for normal development. The essential nature of these genes suggested that the more plausible route to study the role of SUMO in plant development is to study the nature of interactions of SUMO and plant proteins.
SUMO interacts with proteins in two ways. Covalent modification of proteins by SUMO involves the formation of an isopeptide bond between a receptor lysine residue in the target protein and the C-terminal glycine residue of SUMO.Citation6 This follows an ATP-dependent E1→E2→E3 reaction. Proteins covalently modified by SUMO are traditionally called SUMO targets or substrates. SUMO also interacts with proteins non-covalently.Citation7 In the cases studied, SUMO already covalently attached to proteins, recruits other regulatory proteins that recognize SUMO through a SUMO-Interaction Motif (SIM). These proteins are hence described as SUMO-Interacting Proteins (SIPs). A large number of putative SUMO targets were identified in Arabidopsis using different approaches.Citation8-Citation11 These proteins are involved in a variety of cellular processes and developmental programs; however, they are specifically enriched in nuclear proteins, transcription factors, and proteins involved in RNA metabolism. Regulation of abiotic stress responses seems to be a major biological program controlled by SUMO as seen previously by the accumulation of SUMO~protein conjugates in response to stress as well as by studying the dynamics of SUMOylation of specific targets during stress.Citation2,Citation12 On the other hand, several SIPs were identified in yeast and mammals through serendipity, crystal structure analysis of a protein that interacts with SUMO both covalently and non-covalently, and through limited screens.Citation13-Citation17 These findings suggested that non-covalent interactions of SUMO contribute significantly to the role of SUMO in cellular and organismal development. However, a large-scale screen to identify SIPs in a higher multicellular eukaryote was lacking.
In our recent paper,Citation18 we performed a large-scale yeast two-hybrid screen to identify Arabidopsis SUMO-Interacting Proteins. We used three SUMO isoforms (SUMO1, 2, and 3) as baits in three different screens against whole-plant yeast two-hybrid libraries. We sequenced the Arabidopsis cDNA inserts present in more than 750 yeast colonies, and to our surprise we found that 15 proteins interacted with the different SUMO isoforms repeatedly. The SUMO conjugating enzyme (SCE) and E3 ligase (SIZ1) were among these SIPs, as well as 12 proteins that belong to two groups. The first group of SIPs spans six histone and DNA methyltransferases and demethylases that regulate chromatin methylation state by the reversible attachment and removal of methyl groups to and from histones and DNA, whereas the second group of proteins comprises six RING-type Ub E3 ligases that we showed to be SUMO-Targeted Ub E3 ligases (STUbLs). The yeast-two hybrid clones for all these proteins contained regions of the proteins that spanned at least one putative SIM, and we showed that the interaction between SUMO1 and one of these proteins (AT-STUbL1) depends on the SIM. Additionally, the SIM is also required for proper subnuclear localization and for the function of one of the STUbLs that we tested in more detail (AT-STUbL4). The repeated isolation of these proteins suggests that their interaction with SUMO must be physiologically relevant. It also implicates SUMO in the developmental programs they participate in. For example, the isolation of six histone and DNA methyltransferases and demethylases implicates SUMO in the regulation of chromatin methylation state. Interestingly also, these proteins perform reversible functions and have been implicated in important developmental transitions. SDG8, for instance, is a histone methyltransferase that contains a SET (Su(var)3–9, E(z), and Trithorax) domain which is a signature motif for histone methyltransferases that are involved in the epigenetic modification of chromatin.Citation19 SDG8/EFS/CCR1 was first identified as an early flowering mutant,Citation20 and subsequently found to mediate di- and tri-methylation of histone H3 at K36 (H3K36) at FLOWERING LOCUS C (FLC) leading to the activation of FLC expression and a delay of flowering.Citation21,Citation22 sdg8 mutants also exhibit global reduction in the levels of tri-methylation at H3K9 and H3K36 and di-methylation at H3K36, and produce more shoot branching than wild type.Citation23 Additionally, sdg8 was found to be allelic to ccr1 (carotenoid chloroplast regulatory 1), to have altered carotenoid composition and reduced tri-methylation and increased di-methylation at H3K4 surrounding the translation start site of the CAROTENOID ISOMERASE (CRTISO) gene.Citation24 The interaction of SUMO and SDG8 might hence be involved in the regulation of its activity in some or all of these processes. On the other hand, a Jumonji (JMJ)-family protein also interacted with SUMO repeatedly in the yeast two-hybrid. Substantial evidence suggests that JMJ family proteins act as histone demethylases, and that the JmjC domain is involved in this activity. ELF6, REFCitation6 and AtJMJ4, for example, are JMJ-family proteins that regulate flowering time and brassinosteroid signaling by modulating the H3k9 and H3K4 methylation status of target genes.Citation25-Citation27 It is curious that the Arabidopsis SUMO-Interacting Proteins include two groups of proteins of contrasting functions, possibly regulating the expression of common target genes in a dynamic and antagonistic manner.Citation28 For example, the Arabidopsis FLC locus, which is heavily regulated by methylation, is targeted by SDG8, SDG25, and REFCitation6.Citation21,Citation22,Citation25 It is conceivable that SUMO acts as a switch that regulates the recruitment of these chromatin modifiers to chromatin ().
Figure 1. SUMO-Interacting Proteins regulate protein activity and stability. A hypothetical transcription factor is SUMOylated, which enhances its transcriptional factor activity or renders it repressive functions (top). SUMO, attached to this transcription factor, could also recruits other proteins such as chromatin remodeling enzymes that may regulate chromatin methylation state (bottom right), or a polySUMO chain can form and this recruits SUMO-Targetd Ub E3 ligases that mediate the turnover of these transcription factors through the 26S proteasome (bottom left).
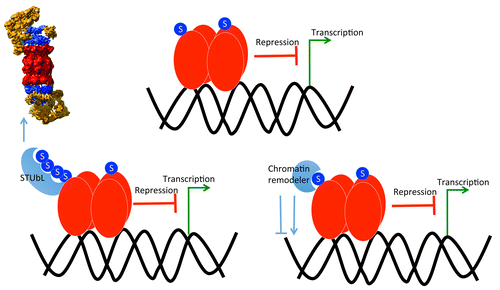
The second group of SUMO-Interacting Proteins includes six Ub E3 ligases. Prior evidence has established that some Ub E3 ligases recognize and target polySUMOylated proteins for degradation by the 26S proteasomeCitation16,Citation17,Citation29 (), and hence these ligases are called SUMO-Targeted Ub E3 Ligases (STUbLs). This form of regulated protein turnover that is stimulated by polySUMO chains represents crosstalk between the SUMO and Ub systems where the degradation of SUMOylated proteins is an alternative route to deconjugation by SUMO proteases. Functionally, these proteins are implicated in the maintenance of genome integrity in yeast and mammals, and loss of activity of the mouse STUbL RNF4 leads to embryonic lethality.Citation30-Citation33 We provided evidence that the six SUMO-Interacting Ub ligases are likely the Arabidopsis STUbLs (AT-STUbLs). They interacted with SUMO1 and/or SUMO2 in the yeast two-hybrid, they contain multiple putative SIMs and a RING domain characteristic of STUbLs in other organisms, and they are evolutionarily related to STUbLs in mammals and fission and budding yeast. Importantly, the six AT-STUbLs complemented to varying degrees the growth defect phenotype of the fission yeast STUbL mutant rfp1/rfp2, suggesting that they exhibit STUbL activity. Two of these proteins (AT-STUbL1 and AT-STUbL3), and to a lesser extent AT-STUbL6, also complemented the genome integrity phenotype associated with rfp1/rfp2, suggesting that these AT-STUbLs may also have maintained a role in genome stability like their yeast and mammalian homologs. Consistent with roles in the nucleus, all AT-STUbLs localize to the nucleus, and mutations of the SIMs or the RING domains affect subnuclear localization. For example, AT-STUbL4, which localizes to the nucleoplasm, the nucleolus, and to discrete nuclear speckles, requires intact SIM and RING domains for speckle localization or formation.
A survey of possible phenotypes associated with AT-STUbLs T-DNA insertion mutants grown under normal growth conditions indicated that mutations in AT-STUbL4 delay flowering especially in long day growth conditions. This is consistent with a role of AT-STUbL4 in the promotion of flowering. This was confirmed in transgenic plants that overexpress AT-STUbL4, which flowered earlier than wild-type plants, as well as by complementing the at-stubl4 mutant using a transgene that expresses AT-STUbL4 from the 35S promoter. This complementation of the late flowering phenotype of at-stubl4 requires both the SIM and RING domains of AT-STUbL4 since mutations of these domains failed to rescue the late flowering phenotype. We identified Cycling Dof Factor 2 (CDF2) as a target of AT-STUbL4 through yeast two-hybrid assays using AT-STUbL4 as bait. This was confirmed by analyses of expression patterns of flowering time genes that control photoperiodic flowering. CDF2 is a floral repressor that acts together with other CDF proteins to repress the expression of CONSTANS (CO), the central player in photoperiodic flowering in Arabidopsis.Citation34 Overexpression of CDF2 in the phloem companion cells produces extremely late flowering plants due to strong repression of CO expression.Citation35 We overexpressed AT-STUbL4 (from the 35S promoter) in lines that already overexpressed CDF2 in phloem companion cells. Our hypothesis was that if AT-STUbL4 did indeed act on CDF2, the overexpression of AT-STUbL4 should suppress the late flowering phenotype of the parental CDF2 overexpressor line and reduce the levels of CDF2 protein. We found exactly that.
Our findings implicate SUMO in the regulation of chromatin methylation state and important developmental transitions such as the floral transition. The detailed examination of the mechanisms with which SUMO regulates these activities, and of the functions of the newly identified AT-STUbLs will greatly help elucidate some of the roles of SUMO in plant development.
Acknowledgements
I would like to thank Richard Vierstra for reading this manuscript. Work in the original paper was funded by grants to George Coupland from the Max Planck Society and the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich 635.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8:947 - 56; http://dx.doi.org/10.1038/nrm2293; PMID: 18000527
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 2007; 145:119 - 34; http://dx.doi.org/10.1104/pp.107.102285; PMID: 17644626
- Reeves PH, Murtas G, Dash S, Coupland G. early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 2002; 129:5349 - 61; http://dx.doi.org/10.1242/dev.00113; PMID: 12403707
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 2007; 19:2952 - 66; http://dx.doi.org/10.1105/tpc.106.049981; PMID: 17905899
- Ishida T, Yoshimura M, Miura K, Sugimoto K. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS One 2012; 7:e46897; http://dx.doi.org/10.1371/journal.pone.0046897; PMID: 23056518
- Dohmen RJ. SUMO protein modification. Biochim Biophys Acta 2004; 1695:113-31.
- Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep 2007; 8:550 - 5; http://dx.doi.org/10.1038/sj.embor.7400980; PMID: 17545995
- Budhiraja R, Hermkes R, Müller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol 2009; 149:1529 - 40; http://dx.doi.org/10.1104/pp.108.135053; PMID: 19151129
- Park HC, Choi W, Park HJ, Cheong MS, Koo YD, Shin G, Chung WS, Kim WY, Kim MG, Bressan RA, et al. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol Cells 2011; 32:143 - 51; http://dx.doi.org/10.1007/s10059-011-2297-3; PMID: 21607647
- Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci U S A 2010; 107:17415 - 20; http://dx.doi.org/10.1073/pnas.1005452107; PMID: 20855607
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A 2010; 107:16512 - 7; http://dx.doi.org/10.1073/pnas.1004181107; PMID: 20813957
- Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol Cell Proteomics 2013; 12:449 - 63; http://dx.doi.org/10.1074/mcp.M112.025056; PMID: 23197790
- Takahashi H, Hatakeyama S, Saitoh H, Nakayama KI. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J Biol Chem 2005; 280:5611 - 21; http://dx.doi.org/10.1074/jbc.M408130200; PMID: 15569683
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 2005; 435:979 - 82; http://dx.doi.org/10.1038/nature03634; PMID: 15959518
- Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 2012; 483:59 - 63; http://dx.doi.org/10.1038/nature10883; PMID: 22382979
- Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 2007; 282:34167 - 75; http://dx.doi.org/10.1074/jbc.M706505200; PMID: 17728242
- Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 2007; 282:34176 - 84; http://dx.doi.org/10.1074/jbc.M706025200; PMID: 17848550
- Elrouby N, Bonequi MV, Porri A, Coupland G. Identification of Arabidopsis SUMO-interacting proteins that regulate chromatin activity and developmental transitions. Proc Natl Acad Sci U S A 2013; 110:19956 - 61; http://dx.doi.org/10.1073/pnas.1319985110; PMID: 24255109
- Zhao Z, Shen W-H. Plants contain a high number of proteins showing sequence similarity to the animal SUV39H family of histone methyltransferases. Ann N Y Acad Sci 2004; 1030:661 - 9; http://dx.doi.org/10.1196/annals.1329.077; PMID: 15659850
- Soppe WJ, Bentsink L, Koornneef M. The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana.. Development 1999; 126:4763 - 70; PMID: 10518493
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 2005; 7:1256 - 60; http://dx.doi.org/10.1038/ncb1329; PMID: 16299497
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana.. Mol Cell Biol 2008; 28:1348 - 60; http://dx.doi.org/10.1128/MCB.01607-07; PMID: 18070919
- Dong G, Ma DP, Li J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem Biophys Res Commun 2008; 373:659 - 64; http://dx.doi.org/10.1016/j.bbrc.2008.06.096; PMID: 18602372
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 2009; 21:39 - 53; http://dx.doi.org/10.1105/tpc.108.063131; PMID: 19174535
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 2004; 16:2601 - 13; http://dx.doi.org/10.1105/tpc.104.025353; PMID: 15377760
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci U S A 2008; 105:7618 - 23; http://dx.doi.org/10.1073/pnas.0802254105; PMID: 18467490
- Jeong JH, Song HR, Ko JH, Jeong YM, Kwon YE, Seol JH, Amasino RM, Noh B, Noh YS. Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS One 2009; 4:e8033; http://dx.doi.org/10.1371/journal.pone.0008033; PMID: 19946624
- Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 2008; 50:886 - 96; http://dx.doi.org/10.1111/j.1744-7909.2008.00692.x; PMID: 18713399
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 2008; 10:538 - 46; http://dx.doi.org/10.1038/ncb1716; PMID: 18408734
- Cook CE, Hochstrasser M, Kerscher O. The SUMO-targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle 2009; 8:1080 - 9; http://dx.doi.org/10.4161/cc.8.7.8123; PMID: 19270524
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 2012; 26:1196 - 208; http://dx.doi.org/10.1101/gad.189274.112; PMID: 22661230
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 2007; 26:4089 - 101; http://dx.doi.org/10.1038/sj.emboj.7601838; PMID: 17762865
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 2008; 322:597 - 602; http://dx.doi.org/10.1126/science.1162790; PMID: 18948542
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 2008; 59:573 - 94; http://dx.doi.org/10.1146/annurev.arplant.59.032607.092755; PMID: 18444908
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 2009; 17:75 - 86; http://dx.doi.org/10.1016/j.devcel.2009.06.015; PMID: 19619493
