Abstract
Chitin, a polymer of N-acetyl-D-glucosamine, is a component of the fungal cell wall and is not found in plants. Plant cells are equipped with chitin degrading enzymes to digest fungal cell walls and are capable of perceiving chitin fragments (chitooligosaccharides) released from fungal cell walls during fungal infection. Chitin recognition results in the activation of defense signaling pathways. Although chitin is a well recognized pathogen-associated molecular pattern (PAMPs), little is known about the molecular mechanism of chitin signaling. Recent studies identified a number of critical components in the chitin-elicited signaling pathway including a potential receptor, MAPK cascade and transcription factor network. Interestingly, the chitin signaling pathway overlaps with the phytobacterial flagellin- and EF-Tu-elicited signaling pathways, suggesting that plant cells may perceive different PAMPs from various pathogens via specialized receptors and then utilize a conserved, common downstream pathway to mediate disease resistance. Given the fact that fungal pathogens are major problems in many agricultural systems, research on chitin signaling could have significance to sustainable agriculture and biofuel and biomaterial production.
Addendum to: Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G.A LysM receptor-like kinase plays a critical role in Chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008; In press.
Chitin, a polymer of N-acetyl-D-glucosamine, is an important component of fungal pathogenicity, since fungal pathogens with defects in chitin synthesis are significantly less virulent on the original susceptible hosts.Citation5,Citation24 Although plants lack chitin, they do secrete chitin-degrading enzymes.Citation18,Citation22 During fungal infection, plant cells secrete chitinases that release chitin fragments (chitooligosaccharides or chitin oligomers) from fungal cell walls that can act as an elicitor to induce plant innate immunity against the invading pathogen.Citation3,Citation18,Citation22,Citation23,Citation25 In agreement with this, overexpression of chitinase in plants led to enhanced resistance to fungal pathogens.Citation4,Citation11,Citation18 Furthermore, pretreatment of plants with chitooligosaccharides enhances plant resistance against various pathogens.Citation26,Citation29 Additionally, recent gene expression profiling studies demonstrated that chitooligosaccharides were a potent regulator of plant gene expression.Citation6,Citation20Citation29,Citation30 All this suggests that a chitin perception and signal transduction pathway is present in plants to mediate plant disease resistance.
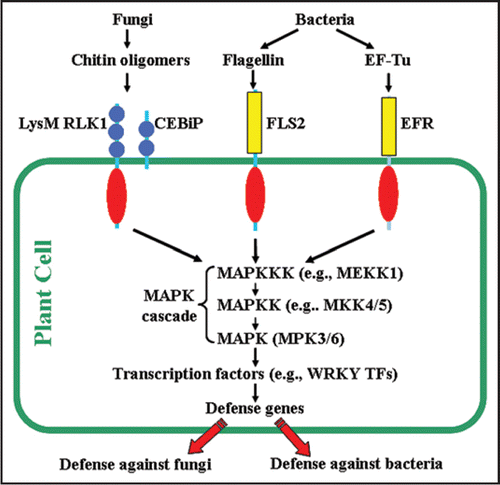
The identification of the chitin receptor(s) complex is pivotal for our understanding of the chitin signaling pathway. Previously, several chitin-binding proteins were detected using biochemical approaches.Citation7,Citation10,Citation17 Recently, Kaku et al.,Citation13 showed that a LysM domain-containing protein CEBiP (chitin elicitor binding protein) binds chitin and plays a critical role in chitin signaling in rice. Since this CEBiP protein does not have an obvious intracellular domain, it very likely needs a partner, such as a receptor-like kinase, to translate the perceived chitin signal into intracellular events. Indeed, very recently, Miya et al.,Citation16 and Wan et al.,Citation29 independently reported that a LysM domain-containing receptor-like kinase 1(LysM RLK1)/chitin-elicitor receptor kinase 1 (CERK1) is critical for chitin signaling in Arabidopsis. Insertional mutations in this gene blocked the induction of virtually all chitin-responsive genes (CRGs) and resulted in greater susceptibility to fungal pathogens. LysM RLK1/CERK1 is a receptor-like kinase with an extracellular LysM domain, which was first identified as a protein module that binds peptidoglycan in bacteria.Citation2,Citation12 Indeed, the legume LysM RLKs, NFR1 and NFR5 in Lotus japonicus, were previously shown to be the putative receptors of Nod factors, which are lipo-chitin molecules essential for nodulation.Citation15,Citation19 Therefore, LysM RLK1/CERK1 very likely plays a critical role in chitin perception, probably through forming a receptor complex with another protein, such as an OsCEBiP-like protein. The Arabidopsis genome harbors three CEBiP-like proteins. We are currently examining the role of each CEBiP-like protein in chitin signaling in Arabidopsis.
Mitogen-activated protein kinases (MAPKs) are important in modulating the response of plant cells to both internal and external stimuli, including pathogen infection.Citation9,Citation27 Increasing data suggest that they may also play an important role in chitin signaling.Citation23,Citation28,Citation30 In particular, MAP kinase 3 and 6 (MPK3/6) were shown to be rapidly activated by chitin in Arabidopsis and their activation depended on upstream MAPK kinases (MKK4 and 5),Citation21,Citation28 suggesting that a MAPK cascade consisting of MKK4/5-MPK3/6 may be involved in chitin signaling.
Transcription factors (TFs) are critical in reprogramming gene expression in plant cells in response to various stimuli. Previous DNA microarray studies suggested plant cells can reprogram gene expression in response to chitin elicitation.Citation6,Citation20 Recently, the use of quantitative transcriptase-polymerase chain reaction (qRT-PCR), in conjunction with DNA microarrays, revealed 118 TF genes responsive to chitin.Citation14 Interestingly, many of these TFs were previously implicated in plant defense (e.g., various WRKY TF genesCitation8). The induction of a number of the chitin responsive TF genes (e.g., WRKY53 and WRKY33) was previously shown to depend on the activation of the aforementioned MAPK cascade,Citation28 suggesting that these TFs may play an important role in regulating other chitin-responsive genes (CRGs). Considering that a large number of TF genes are induced by chitin, one might expect a significant number of plant genes to respond to chitin elicitation. Indeed, the mRNA levels of approximately 900 Arabidopsis genes were shown by DNA microarray analysis to respond to chitin elicitation.Citation20,Citation29 Likewise, in rice, a large number of genes were also shown to be regulated by chitin.Citation6 Consistent with the elicitor role of chitin, many of these regulated genes are defense-related genes, such as those encoding pathogenesis-related proteins, WRKY TFs, and disease resistance proteins (20,29). Furthermore, the knockout of several selected CRGs (e.g., two disease resistance-like protein genes and a putative E3 ligase gene) led to increased susceptibility to the powdery mildew fungal pathogen Erysiphe cichoracearum,Citation20 suggesting a connection between the gene induction by chitin and fungal resistance.
All together, the above findings strongly support that a chitin signaling pathway, as outlined in , is present in plant cells to mediate chitin perception and plant disease resistance. Noteworthy, this pathway is similar to the flagellin and EF-Tu-mediated pathways,Citation1,Citation31 with the major difference in their corresponding receptors. This is also supported by the observation that a large number of genes (441 genes) were commonly upregulated by all three stimuli.Citation29 This similarity suggests that plant cells may have evolved a way to save energy and genetic resources to deal with different pathogens by detecting different PAMPs via different receptors and then employing a similar, conserved pathway to combat different pathogens. Interestingly, the chitin signaling pathway appears to be independent of the pathways mediated by SA, JA and ethylene, at least at the early time points.Citation29,Citation30 However, the detailed relationships, especially at the later time points, between these pathways and those responsive to PAMPs needs to be further investigated.
Abbreviations
| CRGs | = | chitin-responsive genes |
| TF | = | transcription factor |
| LysM RLK1 | = | LysM domain-containing receptor-like kinase 1 |
| CERK1 | = | chitin-elicitor receptor kinase 1 |
| PAMP | = | pathogen-associated molecular pattern |
Figures and Tables
Figure 1 Chitin signaling pathway vs. flagellin-and EF-Tu-mediated pathways. Increasing evidence suggests these pathways share a common, downstream pathway to mediate plant innate immunity against different pathogens, although the initial stages are different, as partially reflected by different receptors. Blue circle: LysM motif, Yellow column: leucine-rich repeats; Red oval: kinase domain.
Addendum to:
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002; 415:977 - 983
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 2000; 299:1113 - 1119
- Boller T. Chemoperception of microbial signals in plant cells. Annu Rev Plant Physiol Plant Mol Biol 1995; 46:114 - 189
- Brogue K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais CJ, Broglie R. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 1991; 254:1194 - 1197
- Bulawa CE, Miller DW, Henry LK, Becke JM. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc Natl Acad Sci USA 1995; 92:10570 - 10574
- Day RB, Akimoto C, Nishizawa Y, Yazaki J, Nakamura K, Fujii F, Shimb K, Yamamoto K, Sakata K, Sasaki T, Kishimoto N, Kikuchi S, Shibuya N, Minami E. Large-scale identification of elicitor-responsive genes in suspension-cultured rice cells by DNA microarray. Plant Biotechnol 2002; 19:153 - 155
- Day RB, Okada M, Ito Y, Tsukada NK, Zaghouani H, Shibuya N, Stacey G. Binding site for chitin oligosaccharides in the soybean plasma membrane. Plant Physiol 2001; 126:1162 - 1173
- Eulgem T. Dissecting the WRKY web of plant defense regulators. PLoS Pathogens 2006; 2:1028 - 1030
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang S, Seguin A, Ellis BE. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends in Plant Science 2006; 11:192 - 198
- Ito Y, Kaku H, Shibuya N. Identification of a high-affinity binding protein for N-acetylchitooligosaccharide elicitor in the plasma membrane of suspension-cultured rice cells by affinity labeling. Plant J 1997; 12:347 - 356
- Jach G, Gornhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Maas C. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J 1995; 8:97 - 109
- Joris B, Englebert S, Chu CP, Kariyama R, Daneo Moore L, Shockman GD, Ghuysen JM. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS MicroBiol Lett 1992; 70:257 - 264
- Kaku H, Nishizawa Y, Ishii Minami N, Akimoto Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 2006; 103:11086 - 11091
- Libault M, Wan J, Czechowski T, Xu D, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin ligase genes involved in plant defense via chitin signaling. Mol Plant-Microbe Int 2007; 20:900 - 911
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003; 425:637 - 640
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 2007; 104:19613 - 19618
- Okada M, Matsumura M, Ito Y, Shibuya N. High-affinity binding proteins for N-acetylchitooligosaccharide elicitor in the plasma membranes from wheat, barley and carrot cells: conserved presence and correlation with the responsiveness to the elicitor. Plant Cell Physiol 2002; 43:505 - 512
- Passarinho P, de Vries SC. Somerville CR, Meyerowitz EM. Arabidopsis Chitinase:a Genomic Survey. The Arabidopsis Book 2002; Rockville MD American Society of Plant Biologists http://www.aspb.org/publications/arabidopsis/ http://dx.doi.org/10.1199/tab.002
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003; 425:585 - 592
- Ramonell K, Berrocal Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S. Loss-offunction mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 2005; 138:1027 - 1036
- Ren D, Yang H, Zhang S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol. Chem 2002; 277:559 - 565
- Schlumbaum A, Mauch F, Vögeli U, Boller T. Plant chitinases are potent fungal growth inhibitors. Nature 1986; 324:365 - 367
- Shibuya N, Minami E. Oligosaccharide signalling for defence responses in plant. Physiol. Mol Plant Pathol 2001; 59:223 - 233
- Soulie MC, Perino C, Piffeteau A, Choquer M, Malfatti P, Cimerman A, Kunz C, Boccara M, Vidal-Cros A. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell Microbiol 2006; 8:1310 - 1321
- Stacey G, Shibuya N. Chitin recognition in rice and legumes. Plant Soil 1997; 194:161 - 169
- Tanabe S, Okada M, Jikumaru Y, Yamane H, Kaku H, Shibuya N, Minami E. Induction of resistance against rice blast fungus in rice plants treated with a potent elicitor, N-acetylchitooligosacchari. Biosci Biotechnol Biochem 2006; 70:1599 - 1605
- Tena G, Asai T, Chiu WL, Sheen J. Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 2001; 4:392 - 400
- Wan J, Zhang S, Stacey G. Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol 2004; 5:125 - 135
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008; 20:471 - 481
- Zhang B, Ramonell K, Somerville S, Stacey G. Characterization of early, chitin-induced gene expression in Arabidopsis. Mol Plant-Microbe Interact 2002; 15:963 - 970
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformatio. Cell 2006; 125:749 - 760