Abstract
The LIM domain is defined as a protein-protein interaction module involved in the regulation of diverse cellular processes including gene expression and cytoskeleton organization. We have recently shown that the tobacco WLIM1, a two LIM domain-containing protein, is able to bind to, stabilize and bundle actin filaments, suggesting that it participates to the regulation of actin cytoskeleton structure and dynamics. In the December issue of the Journal of Biological Chemistry we report a domain analysis that specifically ascribes the actin-related activities of WLIM1 to its two LIM domains. Results suggest that LIM domains function synergistically in the full-length protein to achieve optimal activities. Here we briefly summarize relevant data regarding the actin-related properties/functions of two LIM domain-containing proteins in plants and animals. In addition, we provide further evidence of cooperative effects between LIM domains by transiently expressing a chimeric multicopy WLIM1 protein in BY2 cells.
The LIM domain is a ≈55 amino acid peptide domain that was first identified in 1990 as a common cystein-rich sequence found in the three homeodomain proteins LIN-11, Isl1 and MEC-3. It has since been found in a wide variety of eukaryotic proteins of diverse functions. Animals possess several families of LIM proteins, with members containing 1–5 LIM domains occasionally linked to other catalytic or protein-binding domains such as homeodomain, kinase and SH3 domains. In contrast, plants only possess two distinct sets of LIM proteins. One is plant-specific and has not been functionally characterized yet. The other one comprises proteins that exhibit the same overall structure as the animal cystein rich proteins (CRPs), i.e., two very similar LIM domains separated by a ≈50 amino acid-long interLIM domain and a relatively short and variable C-terminal domain (). The mouse CRP2 protein was the first CRP reported to interact directly with actin filaments (AF) and to stabilize the latter.Citation1 Identical observations were subsequently described for the chicken CRP1 and tobacco WLIM1 proteins.Citation2,Citation3 In addition, these two proteins were shown to arrange AF into cables both in vitro and in vivo and thus join the list of actin bundlers.
To identify the peptide domains of WLIM1 responsible for its actin-related properties/activities, we generated domain-deleted and single domain variants and submitted them to a series of in vivo and in vitro assays.Citation4 Localization experiments established that both LIM domains are required to efficiently target the actin cytoskeleton in tobacco BY2 cells. High-speed (200,000 g) cosedimentation data confirmed that the actin-binding activity of WLIM1 relies on its LIM domains. Indeed, the deletion of either the first or the second LIM domain respectively resulted in a 5-fold and 10-fold decrease of the protein affinity for AF. Importantly, each single LIM domain was found able to interact with AF in an autonomous manner, although with a reduced affinity compared to the wild-type WLIM1. Low-speed (12,500 g) cosedimentation data and electron microscopy observations revealed that the actin bundling activity of WLIM1 is also triggered by its LIM domains. Surprisingly each single LIM domain was able to bundle AF in an autonomous manner, suggesting that WLIM1 has two discrete actin-bundling sites. However, the bundles induced by the variants containing only one LIM domain, i.e., LIM domain-deleted mutants and single LIM domains, differed from those induced by the full-length WLIM1. They appeared more wavy and loosely packed and formed only at relatively high protein:actin ratios. Together these data suggest that LIM domains are autonomous actin-binding and -bundling modules that function in synergy in wild-type WLIM1 to achieve optimal activities.
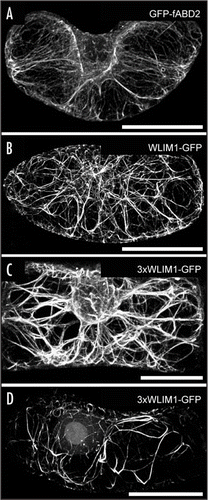
To further assess the mechanism of cooperation between the LIM domains of plant CRP-related proteins, we generated a chimeric protein composed of three WLIM1 copies in tandem (3 × WLIM1, ), and transiently expressed it as a GFP-fusion in tobacco BY2 cells. We anticipated that such a six LIM domain-containing protein displays an even higher actin-bundling activity. () shows the typical actin cytoskeleton pattern in an expanding BY2 cell as visualized using the actin marker GFP-fABD2.Citation5 As previously reported by Sheahan et al.,Citation5 GFP-fABD2 decorated dense, transversely oriented, cortical networks as well as transvacuolar strands connecting the subcortical-perinuclear region to the cortex. Ectopic expression of WLIM1-GFP (BY2 cells normally do not express the WLIM1 gene) induced moderate but perceptible modifications of the actin cytoskeleton structure (). Most AF are arranged in bundles thicker than those observed in GFP-fABD2 expressing cells and fine AF arrays are less frequently observed. As expected, this phenotype was significantly enhanced in cells transformed with the 3xWLIM1-GFP protein (). Indeed, cells were almost devoided of fine AF arrays and exhibited very thick actin cables () that, at times (≈30 %), form atypical long looped structures (). The appearance of such structures may result from the increase of cable stability and thickness induced by the 3xWLIM1-GFP protein, as these parameters are likely to determine, at least partially, the maximal length of actin bundles. Together the present observations support earlier data showing that LIM domains work in concert in LIM proteins to regulate actin bundling in plant cells. Strikingly, vertebrate and plant CRPs invariably contain two LIM domains. The lack, in these organisms, of CRP-related proteins combining more than two LIM domains may be explained by the fact that very thick cables, such as those induced by the artificial 3xWLIM1, may be too stable structures incompatible with the necessary high degree of actin cytoskeleton plasticity. As an exception, a muscle CRP-related protein with five LIM domains (Mlp84B) has been identified in Drosophila.Citation6 However, rather than decorating actin filaments in an homogenous manner, this protein has been found to concentrate in a specialized region of the Z-discs where it stabilizes, in concert with D-titin, muscle sarcomeres.Citation7
The relatively well conserved spacer length (≈50 amino acids) that separates the two LIM domains in vertebrate CRPs and related plant LIM proteins remains an intriguing feature the importance of which in actin cable organization remains to be established. Using electron microscopy we are currently evaluating the effects of the modification of the interLIM domain length on the structural properties of actin cables.
Figures and Tables
Figure 1 Domain maps for wild-type WLIM1 (A) and GFP-fused chimeric 3xWLIM1 (B). A. WLIM1 basically comprises a short N-terminal domain (Nt), two LIM domains (LIM1 and LIM2), an interLIM spacer (IL) and a C-terminal domain (Ct). B. 3xWLIM1 consists of three tandem WLIM1 copies. This chimeric protein has been fused in C-terminus to GFP and transiently expressed in tobacco BY2 cells.
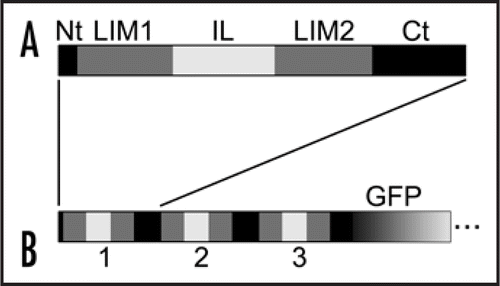
Figure 2 Typical actin cytoskeleton patterns in tobacco BY2 cells that have been transiently transformed, using a particle gun, with GFP-fABD2 (A), WLIM1-GFP (B), and 3xWLIM1-GFP (C and D). For each construct, more than 60 cells were analyzed by confocal microscopy. In the case of 3xWLIM1-GFP, two prevalent patterns have been observed (C and D). Bars = 20 µm.
Addendum to:
References
- Grubinger M, Gimona M. CRP2 is an autonomous actin-binding protein. FEBS Lett 2004; 55:88 - 92
- Tran TC, Singleton C, Fraley TS, Greenwood JA. Cysteine-rich protein 1 (CRP1) regulates actin filament bundling. BMC Cell Biol 2005; 6:45
- Thomas C, Hoffmann C, Dieterle M, Van Troys M, Ampe C, Steinmetz A. Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 2006; 18:2194 - 2206
- Thomas C, Moreau F, Dieterle M, Hoffmann C, Gatti S, Hofmann C, Van Troys M, Ampe C, Steinmetz A. The LIM Domains of WLIM1 define a new class of actin bundling modules. J Biol Chem 2007; 282:33599 - 33608
- Sheahan MB, Staiger CJ, Rose RJ, McCurdy DW. A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol 2004; 136:3968 - 3978
- Stronach BE, Siegrist SE, Beckerle MC. Two muscle-specific LIM proteins in Drosophila. J Cell Biol 1996; 134:1179 - 1195
- Clark KA, Bland JM, Beckerle MC. The Drosophila muscle LIM protein, Mlp84B, cooperates with D-titin to maintain muscle structural integrity. J Cell Sci 2007; 120:2066 - 2077