Abstract
AtPHR1 plays a central role in Pi-starvation signaling in Arabidopsis (Arabidopsis thaliana). Two PHR genes were isolated from rice and designated as OsPHR1 and OsPHR2 based on amino acid sequence homology to AtPHR1. Transgenic plants with overexpression and repression of OsPHR1 and OsPHR2, respectively, were used for investigation of roles of the genes in Pi-signaling pathway and Pi homeostasis under Pi-sufficient and deficient conditions. The results showed that both of the genes are involved in the Pi-signaling pathway, while overexpression of OsPHR2 mimics Pi-starvation stress with enhanced root elongation and proliferated root hairs, and results in the excessive accumulation of Pi in shoots under Pi-sufficient conditions. OsPHR2 regulated proliferation of root hair growth and root elongation suggests that OsPHR2 is involved in both systematic and local Pi-signaling pathways.
A regulation system for the Pi-starvation signaling pathway in Arabidopsis has been proposed.Citation1 In this regulation system, PHR1 is sumoylated by an AtSIZ1-dependent process.Citation2 AtSIZ1 is a plant small ubiquitin-like modifier (SUMO) E3 ligase that is a focal controller of Pi starvation-dependent responses. Downstream of AtPHR1, miRNA399 as a PHR1 target is specifically induced by Pi-starvation. miRNA399 reciprocally regulates the gene PHO2/UBC24 which encodes a critical component for maintenance of Pi homeostasis.Citation3–Citation5 AtIPS1 is induced under Pi-starvation conditions,Citation6,Citation7 and it works as an inhibitor of miRNA399, due to its complementarity with miR399 which is interrupted by a mismatched loop at the predicted miR399 cleavage site.Citation3
AtPHR1 is a member of a large gene family consisting of 11 PHR-like genes in Arabidopsis.Citation8 In rice, the monocot cereal model plant, only two genes homolog to AtPHR1 were found. Our research revealed that both OsPHR1 and OsPHR2 are involved in Pi-signaling pathway. Similar with AtPHR1, repression of OsPHR1/2 downregulates the Pi-starvation-inducible genes, including OsIPS1 and OsIPS2, two AtIPS1 homologs in rice.Citation6,Citation7 Although the functions of OsIPS1 and OsIPS2 were not identified, it could be suggested that they may have the same function as AtIPS1 since they also share the 23-nt nucleotide motif that was functional in AtIPS1 which inhibits miR399 activity, but do not have any effect on miRNA accumulation.Citation3,Citation7
Different function of OsPHR1 and OsPHR2 was found. Overexpression of OsPHR2, but not OsPHR1, mimic Pi-starvation signaling under Pi-sufficient conditions indicated by induced expression Pi-starvation-inducible genes, miRNA399 and several Pi transporters. Overexpression of OsPHR2 also enhanced root elongation and proliferated root hair growth, suggesting the involvement of OsPHR2 in Pi dependent root architecture alteration by both systematic and local pathways. OsPHR2 overexpression results in the excessive accumulation of Pi in shoots under Pi-sufficient conditions. Excretion of phosphate-salt from old leaf tips under solution culture with normal Pi-supplement was observed indicting impaired remobilization of Pi in old leaves to young leaves ().
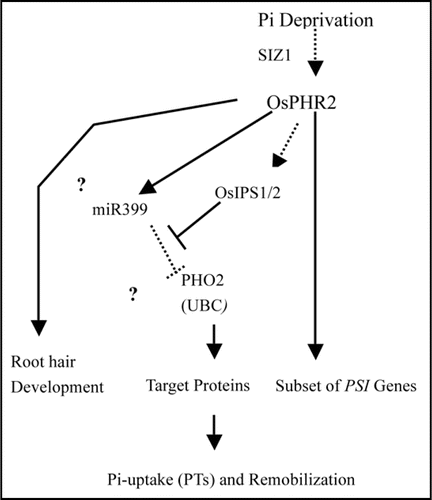
Based on the Pi-starvation-inducible nature of miR399 and its strong repression in a phr1 mutant, miR399 and PHO2 are placed in a branch of the Pi-signaling network downstream of PHR1.Citation9 As for OsPHO2, miR399-mediated transcript degradation was not found under growth condition that was applied; transcription abundance of OsPHO2 was not significantly changed in spite of the change in OsmiR399 levels. Our data suggest that OsPHR2 may positively regulate OsmiR399, and OsPHO2 may not be simply contorted by OsmiR399 in Pi homeostasis in rice; it is also possible that the regulation may affected by other factors such as accumulation of OsIPS1 or controlled by posttranscriptional regulation that needs to be further studied. According to the results, a hypothetical model for OsPHR2 regulated Pi-signaling pathway in rice is proposed in .
Figures and Tables
Acknowledgements
This work was supported by National Basic Research and Development Program of China (2005CB120900) and the Special Program of Rice Functional Genomics of China.
Addendum to:
References
- Schachtman DP, Shin R. Nutrient sensing and signaling: NPKS. Ann Rev Plant Biol 2007; 58:47 - 69
- Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 2005; 102:7760 - 7765
- Franco Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 2007; 39:1033 - 1037
- Chiou TJ, et al. Regulation of phosphate homeostasis by microRNAs in Arabidopsis. Plant Cell 2006; 18:412 - 421
- Fujii H, et al. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 2005; 15:2038 - 2043
- Wasaki J, et al. Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytol 2003; 158:239 - 248
- Hou XL, et al. Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environm 2005; 28:353 - 364
- Todd CD, et al. Transcripts of MYB-like genes respond to phosphorous and nitrogen deprivation in Arabidopsis. Planta 2004; 219:1003 - 1009
- Bari R, et al. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 2006; 141:988 - 999