Abstract
CLE, which is the term for the CLV3/ESR-related gene family, is thought to participate in CLAVATA3-WUSCHEL (CLV3-WUS) and CLV3-WUS-like signaling pathways to regulate meristem activity in plant. Although some CLE genes are expressed in meristems, many CLE genes appear to express in a variety of tissues/cells. Here we report that CLE14 and CLE20 express in various specific tissues/cells outside the shoot/root apical meristem (SAM/RAM), including in highly differentiated cells, and at different developmental stages. Over-expressing CLE14 or CLE20 also causes multiple phenotypes, which is consistent with its expression pattern in Arabidopsis. These results suggest that CLE genes may play multiple roles and involve other signaling cascades in addition to the CLV3-WUS and CLV3-WUS-like pathways.
Intercellular communication and coordination between adjacent cell populations are critical for cell-fate specification, as well as for meristem organization and maintenance. In the shoot apical meristem (SAM), local signaling, which involves the CLAVATA3-WUSCHEL (CLV3-WUS) negative feedback loop, controls stem cell homeostasis and SAM activity.Citation1 As well, it has been suggested that a CLV3-WUS-like negative feedback pathway operates to control root apical meristem (RAM) activity. This view is supported by the facts that a WUS-related homeobox gene, WOX5, is expressed in cells of the quiescent center (QC) in the RAM, and that loss-of-function of WOX5 in the QC leads to the differentiation of the adjacent root cap initials (RCI), whereas gain-of-function blocks the differentiation of derivatives of the RCI in the root.Citation2 Additional support for the function in the RAM of a CLV3-WUS-like pathway, comes from observations that CLE genes (collectively referred to as the CLV3/ESR-relate gene family) are not only expressed in the RAM,Citation3,Citation4 but also, that overexpression of some CLE genes triggers premature termination of the RAM.Citation5 In this regard it has been recently reported that CLE40, which expresses in the differentiating daughter cells of the distal root stem cells, restricts WOX5 expression and promotes differentiation of stem cells in the RAM.Citation6 Taken together these data suggest a CLV3-WUS-like feedback loop acts to negatively regulate RAM activity in plants.
Our previous results have shown that CLE14 and CLE20 express in specific cells of roots, and that overexpression of CLE14 or CLE20 in Arabidopsis triggers early termination of the RAM in a CLAVATA1 (CLV1)-independent, but CLAVATA2 (CLV2)-dependent manner.Citation7,Citation8 We also showed that both CLE14 and CLE20 peptides inhibit, irreversibly, root growth by reducing cell division rates in the RAM.Citation7 CLV2 and CRN (a receptor-like protein kinase, also known as SOL2, isolated as a suppressor of root-specific overexpression of CLE19) are required for CLE14 and CLE20 peptide functions in vitro.Citation9,Citation10 Using computational modeling approaches we further demonstrated that 12-amino-acid CLE14 and CLE20 peptides may function through a potential heterodimer/heterotetramer CLV2-CRN complex.Citation7
CLV3 expresses exclusively in the stem cells of the SAM, and it has been consistently shown that the CLV3 peptide is required for homeostasis of the stem cells and for the maintenance of the SAM.Citation1 Although some CLE genes are found to express in meristems, many CLE genes appear to express in an array of tissues and cells, including highly differentiated tissues/cells.Citation3,Citation4 In this report we show that CLE14 and CLE20 express in specific tissues outside the RAM and SAM of Arabidopsis, including highly differentiated cells, and at different developmental stages. Overexpressing CLE14 or CLE20 also causes multiple phenotypes, which is consistent with its expression pattern in Arabidopsis. These results suggest that CLE genes may play multiple roles in regulating the developmental fate of cells, which includes, but is not limited to, stem cells, and also may be involved in other signaling cascades in addition to the CLV3-WUS pathway.
CLE14 and CLE20 Express in Specific Tissues and Cells Outside the RAM and SAM in Arabidopsis
Using promoter and GFP translational fusion reporter constructs we report that CLE14 expresses specifically in root epidermal cells including the root cap and root hairs (), as well as in the epidermis cells of the hypocotyl (), and in the trichomes () of above-ground tissues. CLE14 is also expressed in anthers () and in pollen (). On the other hand, CLE20 expression is found at the sites of initiation of lateral roots ( and I), and as the root develops, expression progresses into the meristem region of the young lateral roots (), and then spreads sequentially into the protoxylem () and metaxylemCitation4 of the mature roots. In the stem CLE20 also expresses in the protoxylem (). CLE20 expression also occurs sequentially in the flowers, appearing first in the sepals (), then in the petals (), followed by expression in the stamens (). Finally, CLE20 expresses in the inner integuments of the young ovules (), and then in the developing embryos (). CLE20 also expresses in the suspensor cells of the immature embryo (). Moreover, CLE20 expression is detectable in the cotyledons, the leaf primordia, and in the root meristem of 3-day-old young seedling ().
It is also worth noting that CLE14 expression does not appear in either the RAM or SAM, but rather expresses in tissues outside the SAM and RAM, with CLE20 expression mainly confined to tissues outside the SAM and RAM. This result suggests that the endogenous CLE14 or CLE20 peptide may act in other signaling cascades, rather than as part of a CLV3-WUS or CLV3-WUS like pathway. In view of the specific expression patterns of CLE14 and CLE20, these results imply that CLE14 and CLE20 may play rather different roles than in the control of SAM and/or RAM activity in Arabidopsis.
Endogenous CLE14 and CLE20 May Act in a Variety of Tissues/Cells Outside the SAM/RAM and at Different Developmental Stages
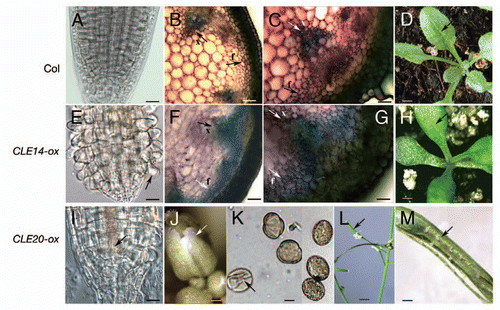
Overexpression of CLE14 or CLE20 triggers premature termination of the RAM in a CLV1-independent, but CLV2-dependent manner in the Arabidopsis root.Citation8 Additionally, our results show that overexpression of CLE14 triggers early differentiation of epidermal cells, leading to root hair development extending into the RAM region in root tip ( and compared to a root tip from a wildtype Col-0 seedling shown in ). Overexpression of CLE14 also causes early lignification of inflorescence stem (, C, F and G). At the top region of the inflorescence stem of the wild-type plant lignin was seen only in xylem cells ( and C), while in plants overexpressing CLE14 lignin staining was strong not only in xylem cells but also in interfascicular fibers ( and G). Moreover, overexpression of CLE14 also leads to a significant reduction in trichome number, and results in dwarf plantsCitation8 with smooth and compact leaves ( and compared to a wild-type Col-0 plant shown in ). These CLE14 overexpression phenotypes are consistent with an expression pattern (), in which CLE14 was found to express in the epidermis of roots and root hairs, and in the epidermal cells of hypocotyl, and in trichomes.
On the other hand, overexpression of CLE20 triggers strong, early differentiation of vascular tissue into the root apex. As shown in , the vascular tissue in root tip uncharacteristically extends through the entire RAM and into the cells immediately adjacent to the QC (). In addition, overexpressing CLE20 also causes small, yellowish floral buds with shortened sepals, petals and stamens (), shrunken pollen (), small siliques () and aborted ovules (). These patterns of CLE20 overexpression are also in agreement with expression patterns (), in which CLE20 expression is found in the xylem () of the mature primary roots, and in the flower organs in a developmental order ().
In summary, we conclude that CLE14 and CLE20 peptides may play roles in regulating cell development in Arabidopsis in different tissues, e.g., epidermis and vascular tissues, outside of the SAM or RAM. Furthermore, based on our modeling resultsCitation7 and the fact that CLV2 and CRN express in a broad range of tissues and cellsCitation10,Citation11 (for example CRN was found to also express in organ primordia and vascular cellsCitation10), we suggest that CLE14 and CLE20 peptides may function in Arabidopsis through the cell signaling pathways involving a CLV2 receptor-like protein and a CRN receptor-like kinase. In view of the expression patterns and overexpression phenotypes of CLE14 and CLE20, and the fact that many of the CLE genes are expressed specifically in multiple cells and tissues outside the SAM and RAM, we propose that endogenous CLE peptides may act in an array of tissues and cells at different developmental stages. Furthermore we suggest that in Arabidopsis, CLE genes may play multiple roles in regulating the development of cells, which include, but are not limited to, stem cells, and also that CLE genes may be involved in other signaling cascades, in addition to the CLV3-WUS and the CLV3-WUS like pathways in the SAM and RAM.
Figures and Tables
Figure 1 The expression patterns of CLE14 and CLE20 analyzed by translational fusion of GFP to the CLE gene; driven by the CLE promoter. (A–G) expression patterns for CLE14. (A, B or C) the root tip (A and B), or root mature region (C) of a 7-day-old transgenic seedling; (D) hypocotyl region of a 7-day-old transgenic seedling; (E–G) GFP signals from a trichome (E), from mature anthers (F) and in a germinating pollen grain from the plant harboring a pCLE14::CLE14-GFP construct. (H–BB) expression patterns for CLE20. (H and I) GFP signals in the lateral root initiation sites of 7-day-old transgenic seedlings; (J) a young lateral root; (K) mature region of a root from a 10-day-old transgenic seedling; (L–N) cross-sections of a 10-day-old transgenic root (L and M) and the inflorescence stem of a 5-week-old transgenic plant (N); (O–AA) CLE 20 expresses in floral organ sepals (O), petals (P), anthers (Q), inner integuments of immature (R and S) and mature (T and U) ovules, developing embryos at the globular stage (V–X), in the suspensor of a embryo at the heart stage (Y), in the mature embryos (Z and AA); (BB) a 3-day-old transgenic seedling. (L, T and V), differential interference contrast (DIC) bright field image; (A, C, H and I) overlay of GFP and DIC images; the remainder are viewed only for GFP. Arrows point to: epidermis in (B and D); a root hair in (C); a trichome in (E); an anther in (F and Q); a pollen tube in (G); lateral root initiation sites (H and I); the RAM in a young lateral root (J); protoxylem of the vascular tissues of the mature roots in (K–M) and stems in (N); a floral organ sepal in (O) and petal in (P); integuments of immature (R and S) and mature (T and U) ovules; suspensor of an embryo at the heart stage in (Y); root meristem, leaf primordia and cotyledons in (BB). Bar = 1 mm in (F, O–Q and BB). Bar = 50 µm for all others. (A–C and H–M) are from Meng et al ().Citation7 The expression analyses of CLE14 and CLE20 were performed as described by Meng et al.Citation7
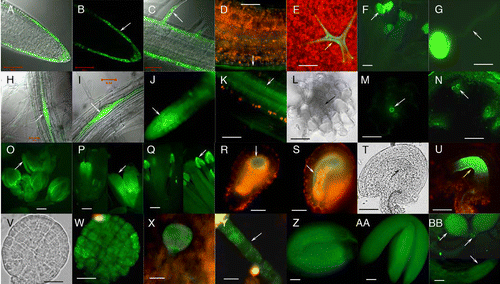
Figure 2 Overexpressing CLE14 or CLE20 causes multiple phenotypes in Arabidopsis plants. (A–D) Non-transgenic wild-type Col-0 plants (Col). (E–H) Transgenic plant overexpressing CLE14 (CLE14-ox). (I–M) Transgenic plant overexpressing CLE20 (CLE20-ox). (A, E and I) the RAM of root tips of the 7-day-old Arabidopsis seedlings. (B, C, F and G) lignification in the inflorescence stems of Arabidopsis. Hand-cut sections of a living inflorescence stem (10–50 µm section from the top part of the stem adjacent to the oldest silique) of 6-week-old plants stained with Toluidine Blue O (0.05% Toluidine blue O in 0.1 M phosphate buffer, pH 7.0). Lignin stains blue-green. (D and H) 21-day-old Arabidopsis plants. (J) an inflorescence tip; (K) pollen; (L) small siliques; (M) an opened small silique. Pollen from plants overexpressing CLE20 is shown to the left, while wild-type pollen (Col-0) is shown to the right in (K). Arrows in (B, C, F and G) point to interfascicular fibers (f) and to xylem (x). Arrows in (D and H) point to a leaf dense with trichomes (D) or to a leaf with fewer trichomes (H), respectively. Arrows in (E and I) point to a root hair (E) or to the vascular tissue (I), respectively. Arrows point to a small and yellowish floral bud with shortened sepals, petals (J), to wrinkled pollen from plants overexpressing CLE20 (K), to a small silique (L) and to an aborted ovule (M). Bar = 20 µm in (A–C, E–G and I); bar = 5 µm in (K); bar = 2 mm in (D and H); bar = 0.5 mm in (J and M); bar = 5 mm in (L). (A, E and I) are from Meng et al.Citation7 Overexpression of CLE14 and CLE20 were performed as described by Meng et al.Citation8
Acknowledgements
We thank Drs. S. Ruzin and D. Schichnes of the UCB Biological Imaging Facility for assistance. This research was supported by a National Science Foundation grant to Lewis J. Feldman and Jennifer C. Fletcher (Arabidopsis 2010 MCB-0313546). Ling Meng was partially supported by an award from the Agricultural and Environmental Chemistry Graduate Program at UC, Berkeley.
Addendum to:
References
- Williams L, Fletcher JC. Stem cell regulation in the Arabidopsis shoot apical meristem. Curr Opin Plant Biol 2005; 8:582 - 586
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007; 446:811 - 814
- Jun JH, Fiume E, Roeder A, Meng L, Sharma K, Osmont KS, et al. Comprehensive analysis of CLE polypeptide signaling gene expression and over-expression activity in Arabidopsis. Plant Physiol 2010; 154:1721 - 1736; PMID: 20884811; http://dx.doi.org/10.1104/pp.110.163683
- Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol 2003; 51:415 - 425
- Strabala TJ, O'donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 2006; 140:1331 - 1344
- Stahl Y, Wink RH, Ingram GC, Simon RA. Signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 2009; 19:909 - 914
- Meng L, Feldman L. CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 2010; 232:1061 - 1074
- Meng L, Ruth KC, Fletcher JC, Feldman L. The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Mol Plant 2010; 3:760 - 772
- Miwa H, Betsuyaku S, Iwamoto K, Kinoshita A, Fukuda H, Sawa S. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol 2008; 49:1752 - 1757
- Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008; 20:934 - 946
- Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of CLAVATA1 receptor-like kinase. Plant Cell 1999; 11:1925 - 1933