Abstract
Xanthomonas campestris pv. vesicatoria (Xcv) is the causal agent of bacterial spot disease in tomato (Solanum lycopersicum) plants. We recently identified a MAPKKK gene, SlMAPKKKε, which is required for tomato resistance to Xcv strains and encodes a positive regulator of cell death. We also provided evidence that the MEK2 MAPKK, and the WIPK and SIPK MAPKs act downstream to MAPKKKε in Nicotiana benthamiana plants. Here, we used the virus-induced gene silencing technique to assess whether tomato homologs of MEK2 (SlMKK2), SIPK (SlMPK1 and SlMPK2), WIPK (SlMPK3), and other components of MAP kinase cascades (SlNPK1, SlMEK1 and SlNTF6), which were previously implicated in plant immunity, are involved in disease resistance to Xcv. Silencing of none of the tested genes caused the appearance of disease symptoms in tomato leaves challenged with an avirulent Xcv strain. However, bacterial populations were significantly higher in leaves of plants silenced for SlMKK2 and SlMPK2, as compared to control plants, suggesting that these two genes contribute to disease resistance to Xcv. It remains to be established whether SlMKK2 and SlMPK2 are activated by SlMAPKKKε directly or through a distinct MAPKKK.
A first line of immune responses is activated in plants by transmembrane pattern-recognition receptors (PRRs) upon perception of highly conserved molecular structures of pathogenic microbes, generally referred to as pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs).Citation1 A second line of immune responses, which include a rapid and localized cell death, known as the hypersensitive response (HR), is triggered by plant resistance (R) proteins that specifically recognize microbial effector proteins.Citation2
MAP kinase cascades are key signaling pathways that participate in the activation of immune responses mediated by both PRR and R proteins.Citation3 MAPK modules have been partially delineated to immune responses activated downstream of certain R proteins.Citation4 For example, the tobacco NtSIPK and NtWIPK MAPKs and their upstream NtMEK2 MAPKK were demonstrated by virus-induced gene silencing (VIGS) experiments to be required for resistance to tobacco mosaic virus (TMV).Citation5 Similarly, silencing of the NtNPK1 MAPKKK, the NtMEK1 MAPKK, and the Ntf6 MAPK, which are considered as components of a distinct MAPK cascade, compromised tobacco resistance to TMV.Citation6,Citation7 In tomato plants resistant to Pseudomonas syringae pv. tomato (Pst), two MAP kinase cascades were proposed to act downstream of the R protein Pto based on genetic and biochemical evidence.Citation8–Citation10 The first pathway involves the SlMAPKKKα MAPKKK, the SlMKK2 (similar to NtMEK2) MAPKK, and the SlMPK2 (similar to NtSIPK) and SlMPK3 (similar to NtWIPK) MAPKs, while the second cascade is composed of the SlMKK3 (similar to NtMEK1) MAPKK and a tomato MAPK similar to Ntf6.
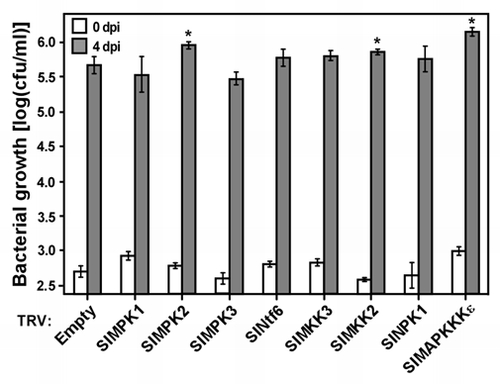
The Gram-negative bacterium Xanthomonas campestris pv. vesicatoria (Xcv) is the causal agent of bacterial spot disease in tomato and pepper (Capsicum annuum).Citation11 Several sources of resistance to Xcv have been identified in tomato lines that specifically recognize Xcv effector proteins and mount efficient immune responses.Citation11 However, signaling components involved in tomato resistance to Xcv are largely unknown. We recently identified a tomato MAPKKK gene, SlMAPKKKε, which is required for HR cell death and disease resistance to Xcv and Pst strains.Citation12 In addition, we showed that MEK2, WIPK and SIPK are required for the cell death triggered by SlMAPKKKε overexpression in N. benthamiana plants.Citation12 Here we tested whether tomato genes encoding homologs of MEK2 (SlMKK2), WIPK (SlMPK3) and SIPK (SlMPK1 and SlMPK2), and other components of MAP kinase modules previously implicated in plant immunity (SlNPK1, SlMEK1 and SlNtf6), are required for disease resistance to Xcv. In these experiments we used a tobacco rattle virus (TRV) vectorCitation13 to silence the genes of interest in plants of the tomato line Hawaii 7981, which is resistant to Xcv bacterial strains expressing the avrXv3 effector gene.Citation14 The efficiency of gene silencing was assessed by quantitative RT-PCR analysis that measured the expression of each target gene in the silenced plants relative to control plants. This analysis indicated a decrease of at least 60% in transcript abundance of each gene targeted by VIGS (). Four weeks after TRV infection, silenced plants were vacuum-infiltrated with a suspension (5 × 104 cfu/ml) of Xcv T2 bacteria expressing the avrXv3 effector gene and monitored in the subsequent days for the appearance of disease symptoms and bacterial growth. Silencing of none of the tested genes, except for the positive control SlMAPKKKε, resulted in the appearance of disease symptoms (data not shown). However, 4 days post-inoculation bacterial populations were slightly, but significantly (p ≤ 0.05) higher in leaves of plants silenced for SlMKK2 and SlMPK2 compared to negative control plants infected with an empty TRV (). Silencing of all other tested genes did not significantly affect bacterial growth. These results suggest that SlMKK2 and SlMPK2 contribute to tomato disease resistance to Xcv. A role for the other tested genes in Xcv resistance cannot be excluded because of either possible functional redundancy with other genes or residual transcript levels in the silenced plants.
Our results provide further evidence for the important function that SlMKK2, SlMPK2, and their homologs from other plant species play in plant immunity. In fact, these kinases have been previously shown to be involved in disease resistance to several pathogens and in PAMP-triggered immunity induced by bacterial flagellin.Citation5,Citation15,Citation16 SlMKK2 and SlMPK2, along with SlMAPKKKε,Citation12 are the first signaling components to be shown to contribute to Xcv resistance. Our previous finding that silencing of either MEK2 or SIPK (homolog of SlMPK2) in N. benthamiana plants attenuates cell death triggered by SlMAPKKKεCitation12 suggests that in tomato SlMKK2 and SlMPK2 also act downstream of SlMAPKKKε. However, immunoprecipitation and phosphorylation assays are required to establish whether these kinases are part of the same MAP kinase module or components of interconnected cascades. Finally, the partial breakdown of resistance observed as a result of SlMKK2 and SlMPK2 silencing suggests the possibility that multiple signaling pathways contribute with an additive effect to the onset of tomato disease resistance to Xcv.
Abbreviations
| HR | = | hypersensitive response |
| MAPK | = | mitogen-activated protein kinase |
| MAPKK | = | MAPK kinase |
| MAPKKK | = | MAPKK kinase |
| Pst | = | Pseudomonas syringae pv. tomato |
| VIGS | = | virus-induced gene silencing |
| Xcv | = | Xanthomonas campestris pv. tomato |
Figures and Tables
Figure 1 Silencing efficiency of genes encoding selected components of MA PK cascades in Hawaii 7981 tomato plants. Plants were infected with Agrobacterium tumefaciens containing a TRV vector either empty or carrying fragments of the following target genes: SlMPK1, SlMPK2, SlMPK3, SlNtf6, SlMEK1, SlMKK2, SlNPK1 and SlMAPKKKε (oligonucleotides and gene fragments used for silencing are available upon request). Relative expression of each target gene was determined by qRT -PCR in silenced plants relative to plants infected with the empty TR V vector. Values are the average of six independent plants silenced in two independent experiments ±SE.
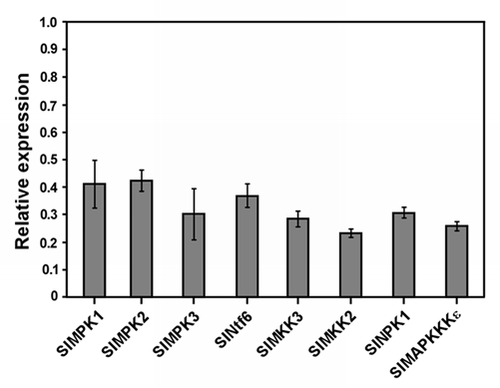
Figure 2 Growth of Xcv T2(avrXv3) bacteria in Hawaii 7981 tomato leaves silenced for genes encoding components of MA P kinase cascades. Leaves of Hawaii 7981 tomato plants silenced for the indicated genes were inoculated by vacuum infiltration with a suspension (5 × 104 cfu/ml) of the avirulent strain Xcv T2(avrXv3). The number of colony-forming units (cfu) per milliliter of plant extract was determined 1 h (white bars) and 4 days (gray bars) post-inoculation (dpi). Bars represent the mean and SE of at least 4 independent experiments, each including at least 5 plants. Asterisks denote statistically significant differences between silenced and control plants infected with empty TR V, as determined by AN OVA and comparisons for all pairs using the Tukey-Kramer honestly significance difference test (p < 0.05).
Acknowledgements
This work was supported by the United States-Israel Binational Agricultural Research and Development Fund (BARD; grant no. IS-4159-08C). S.M. is recipient of an Eshkol fellowship from the Israeli Ministry of Science & Technology.
Addendum to:
References
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 2009; 60:379 - 406
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 2003; 54:23 - 61
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 2010; 61:621 - 649
- Pedley KF, Martin GB. Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 2005; 8:541 - 547
- Jin H, Liu Y, Yang KY, Kim CY, Baker B, Zhang S. Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J 2003; 33:719 - 731
- Jin HL, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang SQ, Staskawicz B, Baker B. NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev Cell 2002; 3:291 - 297
- Liu Y, Schiff M, Dinesh-Kumar SP. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J 2004; 38:800 - 809
- del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 2004; 23:3072 - 3082
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB. Two MAPK cascades, NPR1 and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 2003; 36:905 - 917
- Pedley KF, Martin GB. Molecular basis of Ptomediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol 2003; 41:215 - 243
- Jones JB, Stall RE, Bouzar H. Diversity among xanthomonads pathogenic on pepper and tomato. Ann Rev Phytopathol 1998; 36:41 - 58
- Melech-Bonfil S, Sessa G. Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J 2010; 64:379 - 391
- Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J 2002; 31:777 - 786
- Scott JW, Jones JB, Somodi GC, Stall RE. Screening tomato accessions for resistance to Xanthomonas campestris pv. vesicatoria, race T3. Hortscience 1995; 30:579 - 581
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, et al. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 2002; 415:977 - 983
- Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CD, Nazar RN, Robb J, et al. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol 2009; 150:320 - 332