Abstract
Plant phytohormone, Indole-3-acetic acid (IAA), is synthesized by tryptophan (trp) dependent and independent pathway. Here we report that tryptophan auxotroph mutants completely suppressed the abnormalities of auxin over production mutant, superroot2. SUR2 is considered to modulate Trp dependent pathway, resulting IAA accumulation in Arabidopsis. Tryptophan auxotroph mutants showed hyper-sensitivity to the auxin polar transport inhibitor, NPA, on the phenotype of reduced gravitropism. These results together with the results of histochemical analyses, tryptophan auxotroph mutants seem to have a complete defect in Trp dependent IAA biosynthesis pathway, and it is also suggested that the Trp dependent pathway is responsible for the normal root gravitropism.
Introduction
Auxin regulates many developmental processes in plant, and it has been studied for over a century. Multiple pathways have been proposed on biosynthesis of indole-3-acetic acid (IAA), including tryptophan (Trp) dependent and independent pathway.Citation7 Although bacterial pathways are well known, endogenous plant biosynthetic pathway is still scant. Overexpression of the NIT1 and AAO1, which are suggested to be involved in Trp dependent IAA synthesis pathway, does not cause any auxin-related phenotypes, although nit1 mutant showed resistibility to the effects of the putative auxin precursor indole-3-acetonitrile.Citation10 superroot1, superroot2, and yucca mutants is known to have increased IAA content showing long hypocotyl, epinastic cotyledone, and excess lateral root formation.Citation4,Citation8,Citation16 TAA1/TIR2 is recently isolated to be responsible for indole-3-pyruvic acid (IPA) branch of the auxin biosynthetic pathway.Citation12,Citation15 However, it is not known the IAA deficient mutants. Analyses of the mutants with reduced IAA content would provide us a new insight to the mechanisms of IAA biosynthesis.
Biochemical analysis suggested that tryptophan auxotroph mutants, trp2-1, are defective in the Trp dependent IAA biosynthetic pathway.Citation9 Here we report that trp2-1 and trp3-1 are genetically defective on the Trp dependent IAA synthesis pathway, suppressing the sur2 phenotypes. trp2-1 and trp3-1 also showed significant sensitivity to the auxin transport inhibitor, NPA. Although trp2-1 and trp3-1 showed reduced lateral root formation, its phenotype was suppressed by application of Trp, but not by the Indole. Furthermore, activities of the auxin responsible promoter, DR5 and AtAUX2-11, were reduced in tryptophan auxotroph mutants. These results indicate that Trp dependent IAA biosynthesis in root is responsible for the normal root gravitoropism and lateral root formation.
Results
sur2 mutant showed increased IAA content by modulating the Trp dependent IAA biosynthesis pathway.Citation1,Citation2 To examine whether Trp biosynthesis is responsible for phenotypes of the sur2 mutant or not, we generated a series of double mutants of sur2-1 trp2-1, sur2-1 trp2-8, sur2-1 trp3-1, sur2-1 trp3-100, and sur2-1 trp1-1. sur2-1 mutant produced long hypocotyl, epinastic cotyledone and excess lateral root formation from a hypocotyl (), and the sur2 phenotypes were completely suppressed by trp2-1 mutation, which is known as a strong allele of the trp2 mutant (). In the case of the sur2-1 trp2-8 double mutants, trp2-8 mutation partially suppressed the sur2-1 mutation. sur2-1 trp2-8 double mutants produced long hypocotyl, whereas it did not show epinastic cotyledone and excess lateral root formation from a hypocotyl ( and E). This partial suppression by the trp2-8 mutation is consistent to the fact that the trp2-8 is a weak allele of the trp2 mutant. Similarly, trp3-1 and trp3-100 mutation completely or partially suppress the sur2 phenotypes, respectively (). Further, trp1-1, which is known as a strong allele of the trp1 mutant, could also completely suppress the sur2 phenotype ( and K).
To determine the IAA content of sur2-1 trp2-1 double mutant, we measured it by mass spectrometry (GC-SIM-MS) analyses. As shown in , the average level of three replicate measurements of free IAA in the double mutant of sur2 trp2-1 was completely suppressed to the levels of wild type and trp2-1 mutant. These results indicated that the trp2, trp3 and trp1 mutation is epistatic to the sur2 mutation, suggesting that the Trp dependent IAA biosynthesis pathway is completely intercepted in the trp2-1, trp3-1 and trp1-1 mutants.
We suggested that the Trp auxotroph mutants have a defect in Trp dependent IAA biosynthesis pathway. To better characterize the function of Trp dependent IAA biosynthesis on gravitropism, which is known as an auxin related phenotype, we examined the effect of exogeneously applied auxin transport inhibitor, naphthylphthalamic acid (NPA). When wild-type plant was grown on a medium containing 1 µM NPA, reduced gravitropic response was observed (, B, and D), and the reduction of the gravitropic response in trp2-1 and trp3-1 were significantly enhanced (, , C, E and F), indicating that the trp2-1 and trp3-1 mutants showed hyper sensitivity to the NPA at least on the phenotype of gravitropism.
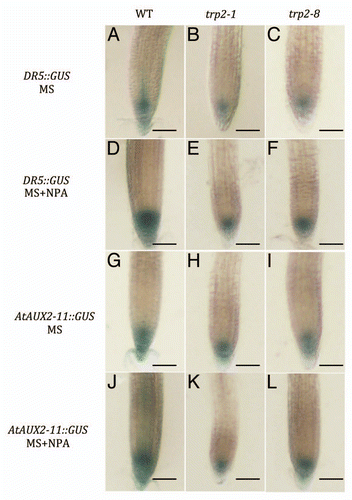
It is known that the DR5::GUS expression is observed at the root tip, and its accumulation is enhanced by an application of the NPA (Casimiro et al. 2001). NPA could block basipetal and acropetal auxin transport, and it is suggested that the enhancement of the GUS activity at the root tip by NPA application was caused by the IAA biosynthesis in vivo. To visualize the auxin spatial accumulation especially at the root tip, we examined the reporter gene activities of DR5::GUS and AtAUX2-11::GUS (Ulmasov et al. 1997, Wyatt et al. 1993). When plants were grown on an agar plate without NPA, similar GUS expression pattern were observed in the wild type, trp2-1, and trp3-1 mutants (, G–I). When they were grown on the agar plates including 1 µM NPA, the GUS activity in the trp2-1 and trp2-8 were significantly reduced in comparison to that of wild type ( and J–L), indicating that the IAA is synthesized via the Trp dependent IAA biosynthesis pathway at the root tip. This hypothesis is consistent to the result that the Trp auxotroph mutants showed hyper sensitivity to the NPA on the gravitropic response. Trp dependent IAA biosynthesis may be responsible for the normal gravitropism.
Discussion
Here we showed that the Trp biosynthesis is responsible for the SUR2 function, and the Trp dependent IAA biosynthesis pathway at the root tip contributes for the appropriate root gravitropism.
SUR2 gene is shown to encode a cytochrome P450 reductase, and SUR2 is shown to catalyze the first committed step in indole glucosinolate biosynthesis by metabolizing indole-3-acetaldoxime to its corresponding aci-nitro compound in vitro. SUR2 is suggested to be a regulator of IAA biosynthesis by regulating the flux of indole-3-acetaldoxime into IAA and/or indole glucosinolate biosynthesis.
Here we showed that trp2-1, trp3-1 and trp1-1 mutation completely suppressed the sur2 phenotypes. This is the first report to show that the Trp dependent IAA biosynthesis is completely intercepted in the loss of function mutant trp2-1, trp3-1. It is known that the trp2-1 and trp3-1 mutants accumulate conjugated types of IAA compared with that of wild type, and it is suggested that the Trp independent IAA biosynthesis pathway is activated in these mutants.
YUCCA and TAA1 are responsible for Trp dependent IAA biosynthetic pathway regulating embryo development.Citation6,Citation12 Together with yucca and taa1 mutants, trp2-1 and trp3-1 mutants would be good materials to analyze the biological function of the Trp dependent IAA biosynthesis pathway.
When plants were grown on agar plate with NPA, enhanced IAA accumulation was observed at root tip, and it would be a result by de novo IAA biosynthesis, not by IAA transport from the shoot. GUS expression levels of the DR5::GUS and AtAUX2-11::GUS were reduced in the trp2-1 and trp3-1 mutants, when plants were grown on agar plates with NPA. This result is consistent to the result that trp2-1 and trp3-1 showed hyper sensitivity to NPA on the root gravitropic response. These results suggested that the Trp dependent IAA biosynthesis pathway mainly contribute to the IAA biosynthesis in the root tissues. As for the shoot tissues, both of the Trp dependent and independent IAA biosynthesis pathways would be activated, and the IAA would be transported from the shoot tissues to root tissues contributing lateral root formation and/or root gravitropisms. This hypothsis is consistent to the result of the inhibition of lateral root formation by the NPA application to the hypocotyl.Citation11
Materials and Methods
Plant material and growth conditions.
Arabidopsis plant seeds were sown on the surface of agar plates with half-strength MS medium, and incubated at 4°C for 3 days. Plants were grown in a laboratory room under continuous illumination of 50–100 µE m−2 s−1 at 22°C. For the chemical application, 1 µM of NPA was added after the autoclave and amino acids, and indole were added before the autoclave.
Quantification of IAA level.
For gas chromatography, single-ion-monitoring mass spectrometry (GC-SIM-MS) analyses, fresh plant material (corresponding to a pool of 10 seedlings grown on agar plates for 10 days in a continuous light condition) was carefully weighed, frozen in liquid nitrogen and stored at −80°C. For extraction of free IAA, the material was ground in liquid nitrogen using a mortar and pestle. After addition of about 0.6 ng of [13C6] IAA (Cambridge Isotope Lab, MS) as an internal standard, the material was extracted in 80% ether and 0.1 mg/ml 2, 6-Di-tret.-butyl-4-methylphenol (BHT) for 60 min. After centrifugation, the supernatant was collected. The pellet was re-extracted again for 90 min, and the supernatant was brought to a water phase in a rotary evaporator. After the pH adjustment to 3.0, the aqueous phase was then partitioned three times against ether containing 0.01 mg/ml BHT. The combined ether phase, which contained ethersoluble acidic and neutral substances, was concentrated until about 800 µl under a nitrogen stream. After the pH adjustment to 10.0 by 1 N NaOH, vortex for 10 min and centrifuge it. Organic phase was discard and repeat this extract twice. After the pH adjustment to 3.0, same volume of ether containing 0.01 g/ml BHT was added. Repeat this extraction twice and dried under a nitrogen stream, and dissolved in methanol. IAA was purified by passing it through a HPLC using a Nucleosil N(CH3)2 column (Senshu, Tokyo, Japan). The mobile phase was 100% methanol and 0.03% acetic acid. Purified solution was dried under a nitrogen stream and trimethylsilylated with MSTFA at 60°C for 15 min.
Splitless injections were made into a GC-SIM-MS system (QP5050A, Shimazu, Tokyo, Japan), equipped with a capillar (DB-1, 0.25 mm i.d. x30 m, film thickness 0.25 µm; J&W Scientific, CA). A linear temperature gradient was applied from 80 to 280°C with a increase of 20°C/min. The injection temperature of the GC was 250°C, the ion source temperature of the MS was 250°C, and a helium flow of 1.2 ml/min. was applied. The ionization potential was 70 electron-volt, and the scan time was 0.2 sec. The percentages of molecules of IAA labeled with 13C were calculated from the relative intensities of m/z 202 to 208, and 319 to 325 ions after subtraction of background. Peak-area ratios for ion pairs were corrected for the abundance of natural isotopes.
Histochemical β-galactosidase assay.
Plant tissue was fixed with 1% glutaraldehyde for 6 h at room temperature. The fixative was then removed and replaced with 1 mg/ml X-Gluc. Tissues were incubated at 37°C for 3 h. The materials were soaked in 95% ethanol and observed by Leica M420 microscope and photographed by Olympus DP50 digital camera.
Note
Grant-in Aid for Creative Scientific Research; Grant-in-Aid for Young Scientists S (19677001) from Japan Society of the Promotion of Science; Grant-in-Aid for Scientific Research for Priority Areas from the Ministry of Education, Culture, Sports, Science, Technology (20061004). Y.M. performed this study. S.S. participated in discussion and helped to draft the manuscript. All the authors read and approved the final manuscript.
Figures and Tables
Figure 1 Genetic analyses of sur2 and trp mutants. (A) sur2-1 (B) trp2-1. (C) sur2-1 trp2-1 double mutant. (D) trp2-8. (E) sur2-1 trp2-8 double mutant. (F) trp3-1. (G) sur2-1 trp3-1 double mutant. (H) trp3-100. (I) sur2-1 trp3-100 double mutant. (J) trp1-1 (K) sur2-1 trp1-1 double mutant. Seedlings were grown on an agar medium for 10 days after germination in the continuous light condition. Bars; 7 mm (A), 3 mm (B–D and F–H), 5 mm (E and I), 1.5 mm (J and K).

Figure 2 IAA contents of Col, sur2, trp2-1 and sur2 trp2-1 double mutant. Seedlings were grown on an agar medium for 5 days after germination. Mean value and standard deviation was calculated by three independent experiments.
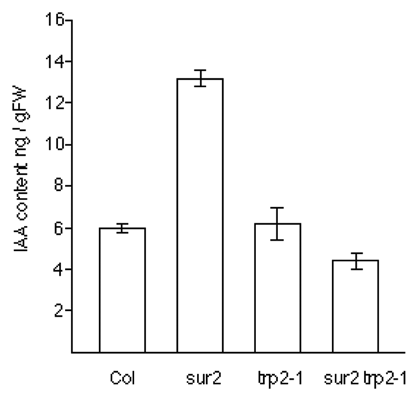
Figure 3 trp mutants showed remarkable sensitivity to NPA. (A and B) wild type. (C and D) trp2-1. (E and F) trp3-1. (A, C and E) Mock treatment. (B, D and F) 1µM NPA treatment. After the incubation for 7 days on agar plates in a vertical position, the plates were rotated 90 degrees and incubate for 2 days. Bars; 1 cm (A and B), 5 mm (C–F).
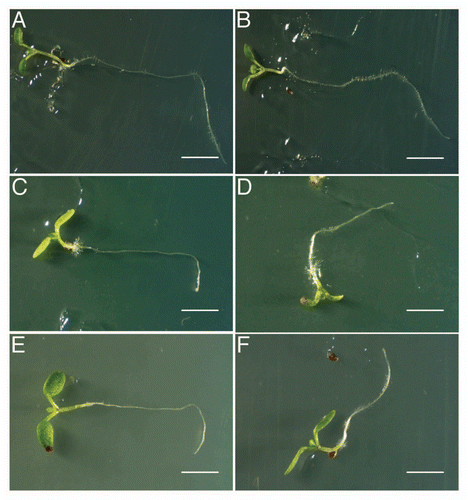
Figure 4 Wild-type (Col), trp2 and trp3 were germinated on MS agar plate (A–C), or a agar plate containing 1 µM NPA (D–F). The orientation of root growth of 30 seedlings was measured after 7 days. Each root orientation was assigned to one of twelve 30 degree sectors. The length of each bar represents the percentage of seedlings showing direction of root growth within that sector.
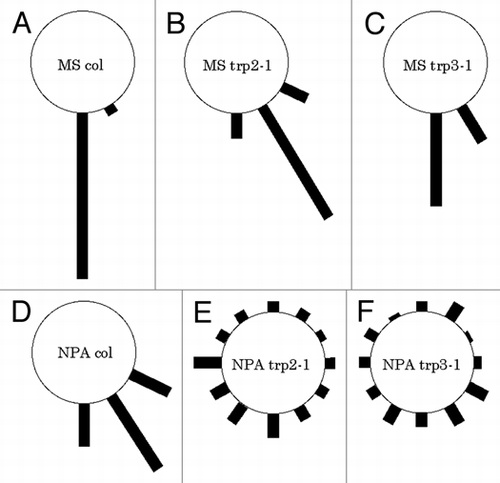
Figure 5 Histochemical staining for GUS activity at the region of root tip in the DR5::GUS (A–F) and AtAUX2-11::GUS (G–L) transgenic plants. Wild type (A, D, G and J), trp2-1 (B, E, H and K), and trp 2–8 (C, F, I and L). Teedlings were on MS agar medium (A–C and G–I) or on agar medium containing 1 µM NPA (D–F and J–L) for 5 days and stained with 2 mg/ml X-Gluc. Bars; 100 µm.
Acknowledgments
We thank to Dr. Masao Tasaka, Dr. Tom Guilfoyle and Arabidopsis Biological Resource Center at OHIO University for providing us AtAux2-11::GUS, DR5::GUS transgenic plants and some plant materials. We also thank Mr. Kensuke Yamazaki for technical assistant.
References
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, et al. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 2000; 97:14819 - 14824
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen DW. CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 2001; 13:101 - 111
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, et al. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 1995; 7:1405 - 1419
- Casimiro I, Marchant A, Rishikesh PB, Beeckman T, Dhooge S, Swarup R, et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 2001; 13:843 - 852
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007; 19:2430 - 2439
- Normanly J, Bartel B. Redundancy as a way of life-IAA metabolism. Plant Cell Physiol 2001; 2:207 - 213
- Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 1998; 14:603 - 611
- Normanly J, Cohen JD, Fink GR. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA 1993; 90:10355 - 10359
- Normanly J, Grisafi P, Fink GR, Bartel B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 1997; 9:1781 - 1790
- Reed RC, Brady SR, Muday GK. Inhibition of auxin movement from the shoot into the root inhivits lateral root development in Aarbidopsis. Plant Physiol 1998; 118:1369 - 1378
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente L, Xie DY, Dole zal K, et al. TAA1-Mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008; 133:177 - 191
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997; 9:1963 - 1971
- Wyatt RE, Ainley WM, Nagao RT, Conner TW, Key JL. Expression of the Arabidopsis AtAux2-11 auxinresponsive gene in transgenic plants. Plant Mol Biol 1993; 22:731 - 749
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiology 2009; 151:168 - 179
- Zhao Y, Christensen SK, Frankhauser C, Cashman JR, Choen JD, Weigel D, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001; 291:306 - 309