Abstract
Organelle DNA in plastids and mitochondria is present in multiple copies and undergoes degradation developmentally. For example, organelle DNA that is detectable cytologically using DNA-fluorescent dye disappears during pollen development. Nevertheless, nucleases involved in this degradation process remain unknown. Our recent study identified the organelle nuclease, DPD1, which has Mg2+-dependent exonuclease activity in vitro. The discovery of DPD1 emerged from Arabidopsis mutant screening and concomitant isolation of dpd1 mutants that retain organelle DNA in mature pollen. DPD1 is conserved only in angiosperms: not in other photosynthetic organisms. Despite these findings, the physiological significance of organelle DNA degradation during pollen development remains unclear because dpd1 exhibits no apparent defects in pollen viability or in the maternal inheritance of organelle DNA. We discuss a possible role of organelle DNA degradation mediated by DPD1, based on a DPD1 expression profile studied using in silico analyses.
Identification of an Organelle Exonuclease DPD1
In plant cells, mitochondria and plastids carry their own genomes that originate from ancestral endosymbionts and which are packed into nucleoid complexes.Citation1,Citation2 Although these organelle genomes encode some (but not all) genes that are necessary for metabolic and energetic functions, they are present in multiple copies, raising the question of whether organelle DNA levels are controlled. Organelle DNAs are regarded as undergoing tissue-specific and/or developmental degradation.Citation3,Citation4 In contrast, the molecular mechanism of organelle DNA degradation (ODD) has remained unclear. We recently identified Defective in Pollen organelle DNA Degradation 1 (DPD1), an organelle exonuclease targeted to both plastids and mitochondria in Arabidopsis.Citation5 This report is the first to describe organelle DNase identified in eukaryotic organisms.
In this work, we specifically investigated the fact that organelle DNAs disappear during pollen development: cytological signals corresponding to organelle DNAs, which are detectable using a fluorescent DNA-binding dye (in our sensitive “pollen squash” method), are undetectable in the mature pollen of most angiosperm species. We performed forward genetic screening in Arabidopsis. The dpd1 mutant was isolated based on its retention of DNA signals corresponding to organelle DNA in mature pollen grains. Characterization of DPD1 and its encoding protein revealed that DPD1 belongs to the exonuclease family and that it is targeted to both plastids and mitochondria. Indeed, DPD1, which is shown to have Mg2+-dependent exonuclease activity in vitro, is highly active in male organs. It is particularly interesting that DPD1 homologs are present in flowering plants but not in moss, green algae and animals. These results implied that, through evolution, angiosperms had acquired the active mechanism of ODD, mediated by DPD1 in the anisogamous male gametophyte.
Remaining Question: Physiological Role of ODD in Pollen
Although they provide evidence, our data leave open the question of the biological significance of this phenomenon: why is organelle DNA degraded in pollen? One possibility we tested in this work is the relevance of degradation to the inheritance mode of organelle DNAs. Given that organelle DNAs are inherited uniparentally (in most cases, including Arabidopsis, from the maternal parent), defective ODD in male organs might alter their inheritance mode. Our genetic analysis, however, demonstrated that dpd1 pollen does not transmit paternal mitochondrial DNAs on the experimental scale we tested (F1 from a cross between Columbia and dpd1, n = 300). Contribution to maternal inheritance is unlikely because ODD proceeds in the pollen vegetative cell that does not fuse to the egg. Considered together, we infer that DPD1 plays non-essential roles in maternal inheritance.
Next, we investigated whether ODD plays a role in pollen viability through nucleotide salvage. For example, multiple copies of mitochondrial DNA are dispensable for mitochondrial function in vegetative and reproductive organs.Citation6,Citation7 Under phosphate-limiting conditions, chloroplast DNA was reportedly degraded in Chlamydomonas, possibly as a compensation mechanism for survival.Citation8 Furthermore, the characterization of plastidial uracil phosphoribosyltransferases showed that plastid functions as the site of uridine nucleotide salvage during early plant development.Citation9 In this work, we were unable to verify this salvage possibility because dpd1 showed no phenotype that results in significant defects in pollen viability. The use of standard in vitro germination medium, however, revealed that dpd1 mutation slightly reduced the germination rate from 60.7% (n = 3) in wild-type plants to 51.0%. Further characterization of dpd1 pollen is necessary to examine whether ODD is involved in pollen germination and other pollen activity.
Using publicly available databases, we examined whether DPD1 expression is associated with stress conditions and/or tissues associated with nutrient salvage. Two interesting facts were revealed. First, the coexpression profiling database ATTED-II (ver. 6.0) revealed that DPD1 expression is associated with Autophagy 7 (ATG7). ATG7 plays a critical role in nutrient recycling and senescence via the autophagy route, which can sequester whole organelles for degradation ( and ).Citation10,Citation11 Carbon starvation and senescence are known to induce expression of a range of autophagy-related genes including ATG7, indirectly suggesting the role of DPD1 in nutrient salvage.Citation12,Citation13 Second, DPD1 expression was examined in the microarray-based expression database Genevestigator.Citation14 As evidenced by our semi-quantitative RT-PCR data, the database revealed that DPD1 transcripts are high in flower and pollen (). Additionally, we found that DPD1 expression is high in senescing leaves and roots, where nutrient salvage is known to be active. A recent report described that root hair and pollen can uptake DNA and can stimulate root and pollen tube growth.Citation15 Taken together with the defective pollen germination rate of dpd1 pollen in vitro, these in silico analyses enable us to propose that ODD in pollen mediated by DPD1 exonuclease is likely to contribute to nutrient salvage for remobilization in developing pollen. In pollen, salvaged nucleotides are important for vegetative cells to support male germ cells in addition to post-pollination development of pollen tubes for fertilization.Citation16
Abbreviations
| ATG7 | = | autophagy 7 |
| DPD1 | = | defective in pollen organelle DNA degradation 1 |
| ODD | = | organelle DNA degradation |
Figures and Tables
Figure 1 Network model of genes coexpressed with DPD1 elucidated by ATTED-II.Citation10
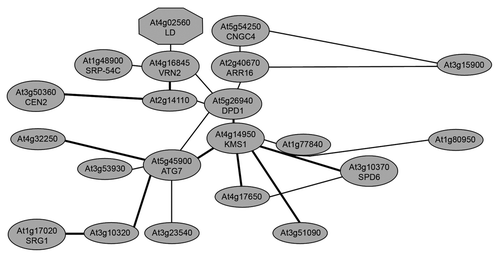
Figure 2 Expression profile of DPD1. Levels of expression are based on signal intensity on high-quality arrays in eight samples from the Genevestigator platform.Citation14 Error bars represent the standard error (S.E.) with the number of arrays used in the calculation.
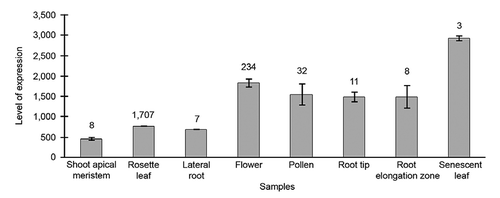
Table 1 Top five genes directly connected with DPD1 based on the coexpressed gene network ATTED-IICitation10
Acknowledgments
This work is supported by Grants-in-Aid for Scientific Research from MEXT (No. 22112516 and No. 22380007 to W.S.) and by the Oohara Foundation (to W.S.).
Addendum to:
References
- Kuroiwa T, Suzuki T, Ogawa K, Kawano S. The chloroplast nucleus: distribution, number, size and shape, and a model for the multiplication of the chloroplast genome during chloroplast development. Plant Cell Physiol 1981; 22:381 - 396
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet 2005; 6:815 - 825; http://dx.doi.org/10.1038/nrg1708
- Sears BB, VanWinkle-Swift K. The Salvage/Turnover/Repair (STOR) model for uniparental inheritance in Chlamydomonas: DNA as a source of sustenance. J Hered 1994; 85:366 - 376
- Sakamoto W, Miyagishima Sy, Jarvis P. Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis Book 2008 (American Society of Plant Biologists)
- Matsushima R, Tang LY, Zhang L, Yamada H, Twell D, Sakamoto W. A conserved, Mg2+-dependent exonuclease degrade organelle DNA during Arabidopsis pollen development. Plant Cell 2011; 23:1608 - 1624; http://dx.doi.org/10.1105/tpc.111.084012
- Wang DY, Zhang Q, Liu Y, Lin ZF, Zhang SX, Sun MX, et al. The levels of male gametic mitochondrial DNA are highly regulated in angiosperms with regard to mitochondrial inheritance. Plant Cell 2010; 22:2402 - 2416; http://dx.doi.org/10.1105/tpc.109.071902
- Preuten T, Cincu E, Fuchs J, Zoschke R, Liere K, Bàrner T. Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J 2010; 64:948 - 959; http://dx.doi.org/10.1111/j.1365-313X.2010.04389.x
- Yehudai-Resheff S, Zimmer SL, Komine Y, Stern DB. Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell 2007; 19:1023 - 1038; http://dx.doi.org/10.1105/tpc.106.045427
- Mainguet SE, GakiÀre B, Majira A, Pelletier S, Bringel F, GuÈrard F, et al. Uracil salvage is necessary for early Arabidopsis development. Plant J 2009; 60:280 - 291; http://dx.doi.org/10.1111/j.1365-313X.2009.03963.x
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 2009; 37:987 - 991; http://dx.doi.org/10.1093/nar/gkn807
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 2002; 277:33105 - 33114; http://dx.doi.org/10.1074/jbc.M204630200
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell 2006; 98:53 - 67; http://dx.doi.org/10.1042/BC20040516
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011; 23:873 - 894; http://dx.doi.org/10.1105/tpc.111.083345
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008; 4207 - 4247
- Paungfoo-Lonhienne C, Lonhienne TGA, Mudge SR, Schenk PM, Christie M, Carroll BJ, et al. DNA is taken up by root hairs and pollen, and stimulates root and pollen tube growth. Plant Physiol 2010; 153:799 - 805; http://dx.doi.org/10.1104/pp.110.154963
- McCormick S. Male gametophyte development. Plant Cell 1993; 5:1265 - 1275