Abstract
The par post-segregational killing locus present on Enterococcus faecalis plasmid pAD1 was the first Type I toxin-antitoxin system described in Gram-positive bacteria. Translation of the 33 amino acid Fst toxin, encoded on RNA I, is suppressed by a 66 nucleotide regulatory RNA, RNA II. RNA I and RNA II are transcribed convergently and interact at dispersed regions of complementarity, establishing a stable complex that accumulates in plasmid-containing cells. RNA II is slowly removed from the complex, allowing translation of RNA I in plasmid-free segregants. Intramolecular structures are also important for regulating translation of RNA I. The Fst toxin contains a putative transmembrane domain and is believed to exert its function at the bacterial cytoplasmic membrane, although its precise target and mode of action have yet to be determined. Numerous chromosomal homologs of pAD1 par have been identified in Gram-positive bacteria suggesting that this locus may play important roles in cellular function.
Introduction: parpAD1 and Its Relatives
The pAD1 par determinant was originally identified as a locus required for maximal stability of the plasmid’s basic replicon.Citation1 The first indication that par might be a toxin-antitoxin (TA) system came from the investigation of a serendipitously isolated pAD1 mini-plasmid that triggered host cell death when induced with cAD1, a peptide pheromone usually required for induction of plasmid conjugation functions.Citation2 Later work showed that this phenomenon resulted from the fortuitous fusion of a pheromone-inducible promoter to the toxin-encoding mRNA of the par locus, RNA I.Citation3,Citation4 Sequence and RNA analysis identified a short transcript convergently transcribed and partially complementary to RNA I,Citation4 designated RNA II. It was later demonstrated that RNA II was capable of counteracting the toxic effects of RNA I both in cis and in trans, confirming its role as the antitoxin of the system.Citation3,Citation5 Toxicity was shown to be due to a 33 amino acid open reading frame designated Fst for faecalis stabilizing toxin.Citation6 It was further demonstrated that the par locus, contained on a fragment of 457 nt, stabilized heterologous plasmids at the expense of host cell growth, confirming its role as a post-segregational killing (PSK) system.Citation3,Citation7,Citation8 More recently, multiple par homologs, sharing both toxin homology and similarity in genetic organization, have been identified on the plasmids and chromosomes of many Gram-positive bacteria.Citation9-Citation11 It is presumed that the plasmid-encoded par homologs perform a function similar to that of pAD1 par, but the function of the chromosomal homologs is as yet unknown. For consistency in nomenclature, we have recommended using either the name of the mobile element or the chromosomal locus designation in subscript with the par componentCitation11 and will use that convention here.
The Genetic Organization of parpAD1 and the Interaction of Its RNAs
The genetic organization of parpAD1 and the structure of its transcripts are shown in and , respectively. The par RNAs are convergently transcribed and share a bidirectional intrinsic terminator. The terminator loop provides one region of complementarity at which the two RNAs interact. The RNAs are also transcribed across a pair of direct repeats, DRa and DRb, in opposite directions which provide a second region of complementarity between RNA IpAD1 and RNA IIpAD1. Interaction at both the terminator loop and the direct repeats is essential for proper regulation of FstpAD1 translation, but the function of these interactions differs. The interaction between the parpAD1 RNAs is initiated at a U-turn motif, originally described for the hok/sok Type I TA system,Citation12 in the terminator loop of RNA IpAD1.Citation13 Mutations in the terminator loop reduce the rate of interaction of the RNAs in vitroCitation13 and abrogate RNA IIpAD1-mediated protection in vivo,Citation5 suggesting that the rate of interaction is important for translational suppression. Following the initial reversible interaction between the terminator loops, binding is rapidly extended to the DRa and DRb repeats, sequestering the initiation codon, interfering with ribosome binding, and inhibiting translation of the toxic peptide, FstpAD1, as determined by ribosomal toeprinting and in vitro translation.Citation6,Citation13
Figure 1. Organization of the pAD1 par locus and the Fst toxin. Converging promoters (black arrowheads labeled P) transcribe the toxin-encoding RNA I (red shaded arrow below line) and the antitoxin RNA II (dark green arrow above line) toward a bi-directional intrinsic transcriptional terminator (converging green arrows). The RNAs are transcribed across direct repeats DRa (pink arrows) and DRb (blue arrows) at which interaction occurs, suppressing translation of the FstpAD1 coding sequence (dark red box on DNA and RNA). The amino acid sequence of the Fst toxin is shown using standard single letter amino acid designations. The essential, conserved hydrophobic domain is shown in bold red print. This forms part of a transmembrane domain in the recently published structure of Fst.Citation22 The two blue underlined amino acids at the N-terminus must be charged to retain toxin function. The non-essential C-terminal tail is shown in green italics.
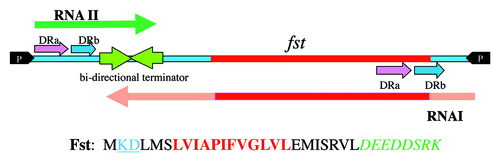
Figure 2. Secondary structures of RNA IpAD1 and RNA IIpAD1. The specific regions of interaction between the RNAs are shaded different colors to coordinate with and labeled accordingly. Interaction is initiated at the U-turn motif (labeled YUNR) present in the loop of the terminator of RNA I (green shaded). This interaction is indicated by the arrow labeled A. The interaction then extends to the direct repeat sequences DRa (pink shaded) and DRb (blue shaded). This interaction is indicated by arrows labeled B and is responsible for preventing translation of FstpAD1, since the initiation codon (I.C.) and the ribosome binding site (SD) are sequestered by the interacting RNAs. The two structures, 5′-SL (blue box) and 5′-UH (red box) are responsible for preventing premature translation of FstpAD1 and RNA IpAD1 stability, respectively.
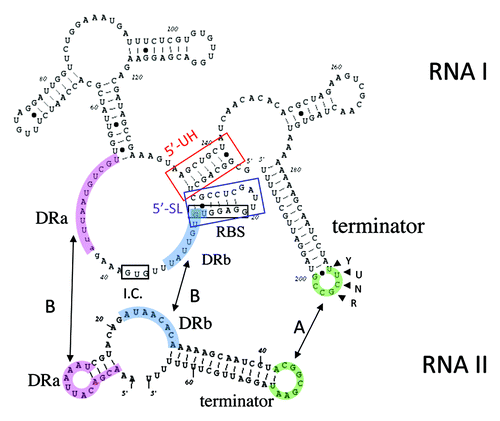
The parpAD1 locus, therefore, has features of both cis- and trans-encoded systems.Citation14 As in cis-encoded systems, the genes for RNA IpAD1 and RNA IIpAD1 overlap providing critical regions of complementarity required for interaction. However, overlap occurs at the 3′ ends of the genes rather than the 5′ ends as in most cis-encoded systems. Like trans-encoded systems, interaction between the parpAD1 RNAs occurs at dispersed regions of complementarity in which different interaction sites play different roles.
Critical Roles of RNA IpAD1 Intramolecular Structures in parpAD1 Regulation
In addition to intermolecular interactions with RNA IIpAD1, intramolecular structures of RNA IpAD1 affect ribosome access to the Shine-Dalgarno (SD) sequence and RNA stability. Two RNA I intramolecular structures, 5′-SL and 5′-UH (boxed and labeled in ), impact parpAD1 function. 5′-SL is a stem-loop that sequesters the FstpAD1 SD, suppressing translation and ribosome binding.Citation6,Citation15 Translational suppression is not complete since low levels of translation can be observed in vitro and wild-type RNA IpAD1 is toxic in vivo in the absence of RNA IIpAD1. However, mutations destabilizing the 5′-SL cannot be established in cells expressing RNA IIpAD1Citation15 in spite of the fact that it is capable of binding to and suppressing translation from such mutants in vitro.Citation6 This discrepancy between in vivo and in vitro results may relate to the timing of RNA IpAD1’s interaction with its two competing partners, ribosomes and RNA IIpAD1. Because the interaction between the RNAs is initiated at the terminator loop, the ribosome binding site of RNA IpAD1 is transcribed and available for ribosome binding before RNA IIpAD1 can initiate binding. The 5′-SL is postulated to temporarily inhibit ribosome binding until the terminator loop can be transcribed. It is also possible that RNA IpAD1 is processed to remove the 5′-SL in vivo before it can be translated, but no such processing product has been observed in spite of multiple attempts.
The 5′-UH is an “upstream helix” composed of the extreme 5′ end of the RNA IpAD1 transcript and a complementary sequence further downstream that folds back to interact with it. This helix sequesters the 5′ nucleotides from digestion by cellular RNases and is at least partially responsible for the greater stability of RNA IpAD1 relative to RNA IIpAD1.Citation16 Mutations in the 5′-UH result in a > 4-fold drop in RNA IpAD1 half-life from > 40 min to around 9 min; the half-life of free RNA IIpAD1 is approximately 4 min. Mutation of the 5′-UH makes RNA IpAD1 more susceptible to RNases J1 and J2, which have 5′-3′-exonuclease activity.Citation17 Whether these are the primary RNases responsible for degradation of RNA IpAD1 is not clear. It is also possible that other features of RNA IpAD1, e.g., its relatively inaccessible 3′ end and its compact structure, may contribute to its stability.
Interaction of parpAD1 RNAs Facilitates the Accumulation of a Stable Complex
In order for Type I TA systems to function as plasmid stabilizing PSK systems, a sufficient pool of the toxin message must accumulate to allow translation after the plasmid has been lost. In the prototypical hok/sok system, this is accomplished by the formation of alternate conformations of the hok mRNA (see x in this issue). In contrast, RNA IpAD1 does not appear to adopt alternate structures that control RNA interaction, degradation, and translation. Rather, interaction of the parpAD1 RNAs leads to stabilization of the RNAs and accumulation of the RNA IpAD1-RNA IIpAD1 complex. In the presence of RNA IpAD1, RNA IIpAD1 basal levels increase more than 2-fold and half-life increases from 4 to 16 min.Citation18 Similarly, the basal level and stability of the RNA IpAD1 destabilizing 5′-UH mutant (see above) was increased more than 2-fold in the presence of RNA IIpAD1.Citation16 These results suggest that formation of the RNA IpAD1-RNA IIpAD1 complex protects both RNAs from degradation by cellular RNases. While most regulatory RNAs appear to destabilize their targets, target stabilization is not without precedent.Citation19 These results led to the following model for regulation of parpAD1 function. Following transcription of RNA IpAD1, the 5′-SL prevents ribosome binding until interaction with RNA IIpAD1 can occur. The translationally inactive complex then accumulates as a pool in the cells with RNA IpAD1 to RNA IIpAD1 ratios maintained at approximately 1:1.1.Citation18 It is possible that the discontinuous nature of the interacting sites in the complex prevents efficient degradation by RNase III which requires at least two helical turns of double stranded RNA for maximal binding and activity.Citation20 The lower stability of RNA IIpAD1 suggests that it is preferentially removed from the complex and degraded by means that have yet to be described. This removal must be active, since in vitro results suggest that the complex does not spontaneously dissociateCitation18 and could involve RNA helicase and/or targeted RNase action. If plasmid remains in the cell, sufficient RNA IIpAD1 is produced to replace that removed from the complex. If the plasmid is lost, degraded RNA IIpAD1 cannot be replaced, the FstpAD1 ribosome binding site becomes accessible, either through the processing of the 5′-SL or by the utilization of a ribosomal standby siteCitation21 (perhaps within the adjacent large unstructured loop), and sufficient toxin is produced to kill the cell.
The FstpAD1 Toxin Is a Small, Probably Membrane Localized, Peptide
The FstpAD1 toxin is a 33 amino acid peptide with a charged N-terminus, a predicted central transmembrane domain, and a highly charged C-terminal tail (). Alanine scanning mutagenesis supplemented with select conservative and non-conservative amino acid changes revealed that the putative transmembrane domain was most important for toxin function.Citation11 Alanine substitutions in many of the amino acids in this region lost toxicity, while substitutions with bulky hydrophobic amino acids leucine and valine were tolerated, consistent with a role in membrane transit. The centrally located P11 residue appears especially important for function since substitution with four different amino acids, including alanine and acidic and basic amino acids, eliminated toxicity. At the N-terminus, substitutions of the two charged amino acids, K2 and D3, with either acidic or basic amino acids retained toxicity. However, alanine substitutions were non-toxic suggesting that polarity at the N-terminus is important for function. Substitution of L4 with either alanine or a charged amino acid eliminated toxicity. In contrast, the charged C-terminal tail appears to contribute little to toxin function. In most cases, alanine substitutions and substitutions reversing amino acid charge had no effect. Indeed, a nonsense mutation at D25 resulting in truncation of the C-terminus was still toxic, indicating that the last eight amino acids are not required for toxicity. A nonsense mutation in the adjacent L24 was non-toxic. It is important to note that the mechanism of testing toxicity could not distinguish degrees of toxicity, so it was not possible to test if toxic mutations might have had reduced toxicity.
An atomic resolution structure of FstpAD1 has been determined in the membrane mimetic dodecylphosphocholine by NMR spectroscopy.Citation22 These results indicated that Fst forms a transmembrane α-helix with the first two and the last seven amino acids protruding. The charged C-terminal seven amino acids are disordered and were predicted to extend from the cytoplasmic side of the membrane. These authors suggested that the primary function of membrane insertion was to facilitate interactions with a specific target rather than being directed against the membrane itself. They also predicted that the disordered C –terminus might become structured upon recognition of the target, but this conclusion conflicts with mutagenic studies indicating that the last eight amino acids are not essential for toxicity.Citation11
Overexpression of FstpAD1 Affects Nucleoid Structure, Segregation and Cell Division
FstpAD1 is toxic to E. faecalis,Citation23,Citation24 S. aureusCitation11 and B. subtilisCitation23 when overexpressed from the native RNA I transcript. Toxicity can also be observed in E. coli if the 5′-SL structure is disrupted.Citation15 In all four species, the primary effect of toxin overexpression is condensation of the nucleoid. In E. coli and B. subtilis this results in elongation of cells, perhaps because the collapsed nucleoid interferes with formation of the division septum at the cell center. In S. aureus, the division septum forms and invaginates but the nucleoid is frequently trapped at the convergence point and completion of cell division is inhibited. E. faecalis cells initially elongate, then produce misplaced division septae and finally mis-segregate the nucleoid producing cells containing little or no DNA. The different effects of Fst may reflect differences in the control of cell division in the different species. In E. coli and B. subtilis nucleoid occlusion systemsCitation25 apparently prevent the formation of division septae over the condensed chromosome. In both S. aureus and E. faecalis nucleoid occlusion appears to be ineffective in stopping invagination of the cell wall or Fst abrogates its function. In S. aureus, new cell wall growth occurs only at the septum,Citation26 so the presence of a condensed nucleoid effectively blocks both division and growth. In the chaining ovococci, however, cell wall growth occurs both longitudinally and septally,Citation27 allowing elongation of Fst-exposed cells with the nucleoid trapped at the division site. In at least some cells, the partition apparatus mobilizes the condensed chromosome, but only into one of the daughter cells.
FstpAD1 Is Active at the Membrane But Its Specific Target Is Unknown
The putative transmembrane domain of FstpAD1 and its importance to toxin function suggest that the peptide is membrane localized. However, exposure to FstpAD1, unlike Hok,Citation28 does not result in the leakage of cell contents and the formation of “ghost cells.” An increase in cell permeability is observed following FstpAD1 overexpression but only after the appearance of cell growth and division anomalies, suggesting that membrane defects may be a secondary effect.Citation23,Citation24 Nisin and Fst have synergistic effects suggesting that they have different but complementary targets.Citation24 Nisin is a pore forming lantibiotic that docks on lipid II and also affects peptidoglycan synthesis.Citation29 Unlike nisin but like Hok, synthetic FstpAD1 has no effect on cell growth when added externally,Citation24 suggesting either that it is modified in some way within the cell or targets a component present only on the inner surface of the membrane or in the cytoplasm. Recent microarray data indicates that overexpression of FstpAD1 results in induction of a variety of energy-dependent membrane transporters; interference with this induction by RNA polymerase mutation or interference of ABC transporter activity with reserpine leads to Fst resistance (Brinkman and Weaver, unpublished). It is possible that hyperactivity of energy-utilizing membrane transporters depletes the cells of energy thereby leading to the observed toxic effect.
parpAD1 Homologs Are Widespread in Gram-Positive Organisms
Work by several groups has revealed that FstpAD1 belongs to a large family of RNA-regulated peptide toxins.Citation9-Citation11,Citation30 These peptides are smaller than 60 amino acids, hydrophobic, and predicted to contain an α-helical transmembrane domain. Indeed, many of the smaller peptides may consist solely of the transmembrane helix. Most are toxic when overexpressed in their native host.Citation30 An exhaustive bioinformatic search across 774 bacterial genomes identified hundreds of these peptides in the γ-proteobacteria and Firmicutes that were divided into eight families.Citation9 FstpAD1 is the founding member of the Fst/Ldr family of peptide toxins, which in this analysis consisted of 161 members. In addition, Kwong et al. reported the identification of more than 200 Fst-related peptides in a diversity of Gram-positive bacteria.Citation10 While there is likely significant overlap between these two lists, it seems apparent that Fst peptides are ubiquitous in Gram-positive bacteria and the related Ldr peptides are prevalent in the γ-proteobacteria. In the Gram-positive bacteria, examination of the DNA sequences surrounding the Fst peptides revealed the existence of all of the elements originally defined in the parpAD1 locus, including the convergent promoters for RNA I and RNA II transcripts, a bi-directional intrinsic terminator, the DRa and DRb interacting sequences, and sequences providing the 5′-SL and 5′-UH of RNA I, suggesting that they may be regulated in a similar manner to parpAD1.Citation10,Citation11 U-turn motifs were not always present in the terminator loop, however, suggesting that some features of the interaction pathway might differ in individual systems. In addition, while the general features of the Fst-encoding par loci are conserved, their sequences are not, particularly in the DRa and DRb regions predicted to be involved in RNA-RNA interaction. This feature would allow related par systems present on different plasmids to operate in the same cell without interfering with one another. Furthermore, a number of par-homologs are chromosomally-encoded. For example, FstEF0409 is present in all sequenced E. faecalis but not E. faecium strains.Citation31 Recent work in our laboratory indicates that it neither interferes with nor is essential for pAD1 par function (Weaver, unpublished results).
Many of the newly identified par homologs are present on plasmids, where they presumably perform the same PSK function as parpAD1.Citation10 Indeed, a par homolog on the well-studied S. aureus plasmid pSK41 has been demonstrated to stabilize a heterologous plasmid and FstpSK41 has been shown to be toxic when overexpressed in E. coli (Kwong and Firth, personal communication). Another abundant class of par homologs is associated with chromosomally-integrated mobile genetic elements including one within the SaPIbov2 staphylococcal pathogenicity island and one phage from Lactobacillus gasseri.Citation10 However, some chromosomally-encoded par homologs appear not to be linked to recognizable mobile genetic elements (MGE). The function of these chromosomally-located par loci, like most other chromosomal TA systems, is unknown. Interestingly, five of the par homologs not associated with MGE are intimately linked to genes involved in carbohydrate metabolism.Citation10,Citation11 This includes parEF0409 located between genes for mannitol phosphotransferase components in E. faecalis, parSSP0870 located between genes for 6-phosphoglucono-lactonase and an aldehyde dehydrogenase in Staphylococcus saprophyticus, parLSEI2682 situated between genes for mannose-6-P isomerase and a two-component signal transduction system in Lactobacillus caseii, a locus in S. aureus MRSA252 located between genes encoding a putative ABC transporter and glycerate kinase, and a Listeria monocytogenes locus downstream of a gene encoding a glycosyl hydrolase. In Streptococcus pneumoniae, a pair of tandemly-encoded par homologs are located between genes for a regulator of a fucose operon and an ABC transporter.Citation9 The locations of these par homologs along with the association of Fst effects on ABC transporters are suggestive of a role in fine-tuning carbohydrate metabolism. This possibility is under active investigation.
Finally, a phylogenetic link between the Fst toxin and the Ldr toxin encoded in the ldr/rdl type I TA system present in the LDR (long direct repeat) sequences in the E. coli K12 genome was recently identifiedCitation9,Citation32 (see also x in this issue for more detail on the ldr/rdl system). A superfamily signature consisting of a transmembrane helix followed by a highly conserved tryptophan with a C-terminal tail of charged amino acids was suggested, though the FstpAD1 prototype lacks the conserved tryptophan. The possible relationship between these peptides is further strengthened by their strikingly similar effects upon overexpression in E. coli. How these apparently related peptides came to reside in such disparate hosts is a mystery. Although Fst homologs are frequently present on mobile genetic elements, phylogenetic analysis showing coherence between the phylogeny of Fst/Ldr peptides and their hosts of origin suggests that their distribution is not due to recent horizontal gene transfer. Interestingly, Ldr expression appears to be regulated by a mechanism similar to that of hok/sok rather than by a par-like mechanism.Citation33 It is interesting to speculate on the evolutionary path that fused an Fst-like peptide to a hok/sok like TA locus.
Conclusion
To date, the hok/sok and par systems remain the best studied Type I TA systems and a comparison of the two systems is instructive. While detailed analysis has identified many similarities between the two systems, there are also significant differences. For example, while both systems utilize regulatory RNAs to control the translation of their respective toxins and U-turn motifs are critical for interaction timing, the Sok RNA is a classic cis-acting antisense RNA while RNA IIpAD1 interacts with its targets via dispersed regions of complementarity in a manner more similar to trans-acting RNA regulators. Both the hok RNA and RNA IpAD1 adopt a compact secondary structure that stabilizes the RNA and suppresses premature translation initiation. However, while the hok RNA structure prevents Sok binding in order to allow a stable pool of the toxin message to accumulate, RNA IpAD1 interaction with RNA IIpAD1 is apparently immediate and it is the complex that accumulates. Both Hok and FstpAD1 are membrane active peptide toxins that must be produced internally to exert their effects. But while Hok expression leads to the production of ghost cells, FstpAD1 causes nucleoid condensation and division inhibition. It seems likely that hok/sok and par evolved independently and found partially convergent means to solve similar problems. The surprising hybrid nature of the ldr/rdl system certainly deserves more attention from an evolutionary perspective.
Significant features of parpAD1 function and regulation remain to be determined. The most pressing issue regarding RNA-mediated regulation is the mechanism of release of RNA IIpAD1-mediated repression of RNA IpAD1 translation. What enzymes and what processes are required to remove RNA IIpAD1 from the RNA complex? Is the 5′-SL of RNA IpAD1 removed or in some other way altered prior to translation? The mechanism of action of the Fst toxins remains unclear. What are the targets on the cytoplasmic membrane to which these peptides bind? How do they insert into the membrane? How does FstpAD1 affect nucleoid structure, partition and division? Why does FstpAD1 have different effects on cell division in different hosts? Examination of this last question may provide insights into the regulation of cell division and chromosomal partition in cocci. The existence of large numbers of par homologs on the chromosomes of Gram-positive bacteria, many unassociated with mobile genetic elements, adds another layer of mystery to these apparently ubiquitous elements. Their association with genes involved in carbohydrate metabolism suggests that they may be responsive to growth conditions. Preliminary work with parEF0409 suggests that both RNAs of this system are regulated in response to growth phase (Weaver, unpublished results). If so, what is their role in regulating bacterial metabolism? Does the RNA II component regulate Fst expression in the same manner in plasmid and chromosomal systems? Do the chromosomal Fst’s have the same target as the plasmid versions? Are the chromosomal versions as lethal as the plasmid versions? Answers to these questions concerning parpAD1 and its chromosomal homologs will go a long way toward determining the evolutionary relationship between them.
| Abbreviations: | ||
| TA | = | toxin-antitoxin |
| PSK | = | post-segregational killing |
| SD | = | Shine-Dalgarno |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I would like to acknowledge the hard work and dedication of post-doctoral fellows, graduate students, research associates, undergraduates, medical students and numerous collaborators over the years that had contributed to the work described herein. This work was funded in part by Public Health Service grant GM55544, National Science Foundation Grant MCB-9723098 and the Division of Basic Biomedical Sciences, Sanford School of Medicine.
References
- Weaver KE, Clewell DB, An F. Identification, characterization, and nucleotide sequence of a region of Enterococcus faecalis pheromone-responsive plasmid pAD1 capable of autonomous replication. J Bacteriol 1993; 175:1900 - 9; PMID: 8384618
- Weaver KE, Clewell DB. Construction of Enterococcus faecalis pAD1 miniplasmids: identification of a minimal pheromone response regulatory region and evaluation of a novel pheromone-dependent growth inhibition. Plasmid 1989; 22:106 - 19; http://dx.doi.org/10.1016/0147-619X(89)90020-6; PMID: 2516332
- Weaver KE, Jensen KD, Colwell A, Sriram SI. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par.. Mol Microbiol 1996; 20:53 - 63; http://dx.doi.org/10.1111/j.1365-2958.1996.tb02488.x; PMID: 8861204
- Weaver KE, Tritle DJ. Identification and characterization of an Enterococcus faecalis plasmid pAD1-encoded stability determinant which produces two small RNA molecules necessary for its function. Plasmid 1994; 32:168 - 81; http://dx.doi.org/10.1006/plas.1994.1053; PMID: 7531349
- Greenfield TJ, Weaver KE. Antisense RNA regulation of the pAD1 par post-segregational killing system requires interaction at the 5′ and 3′ ends of the RNAs. Mol Microbiol 2000; 37:661 - 70; http://dx.doi.org/10.1046/j.1365-2958.2000.02034.x; PMID: 10931359
- Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol Microbiol 2000; 37:652 - 60; http://dx.doi.org/10.1046/j.1365-2958.2000.02035.x; PMID: 10931358
- Weaver KE. Enterococcus faecalis plasmid pAD1 replication and maintenance. Dev Biol Stand 1995; 85:89 - 98; PMID: 8586250
- Weaver KE, Walz KD, Heine MS. Isolation of a derivative of Escherichia coli-Enterococcus faecalis shuttle vector pAM401 temperature sensitive for maintenance in E. faecalis and its use in evaluating the mechanism of pAD1 par-dependent plasmid stabilization. Plasmid 1998; 40:225 - 32; http://dx.doi.org/10.1006/plas.1998.1368; PMID: 9806859
- Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res 2010; 38:3743 - 59; http://dx.doi.org/10.1093/nar/gkq054; PMID: 20156992
- Kwong SM, Jensen SO, Firth N. Prevalence of Fst-like toxin-antitoxin systems. Microbiology 2010; 156:975 - 7, discussion 977; http://dx.doi.org/10.1099/mic.0.038323-0; PMID: 20150240
- Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 2009; 155:2930 - 40; http://dx.doi.org/10.1099/mic.0.030932-0; PMID: 19542006
- Franch T, Petersen M, Wagner EGH, Jacobsen JP, Gerdes K. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol 1999; 294:1115 - 25; http://dx.doi.org/10.1006/jmbi.1999.3306; PMID: 10600370
- Greenfield TJ, Franch T, Gerdes K, Weaver KE. Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol Microbiol 2001; 42:527 - 37; http://dx.doi.org/10.1046/j.1365-2958.2001.02663.x; PMID: 11703673
- Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol 2009; 4:85 - 103; http://dx.doi.org/10.2217/17460913.4.1.85; PMID: 19207102
- Shokeen S, Patel S, Greenfield TJ, Brinkman C, Weaver KE. Translational regulation by an intramolecular stem-loop is required for intermolecular RNA regulation of the par addiction module. J Bacteriol 2008; 190:6076 - 83; http://dx.doi.org/10.1128/JB.00660-08; PMID: 18641135
- Shokeen S, Greenfield TJ, Ehli EA, Rasmussen J, Perrault BE, Weaver KE. An intramolecular upstream helix ensures the stability of a toxin-encoding RNA in Enterococcus faecalis.. J Bacteriol 2009; 191:1528 - 36; http://dx.doi.org/10.1128/JB.01316-08; PMID: 19103923
- Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 2007; 129:681 - 92; http://dx.doi.org/10.1016/j.cell.2007.02.051; PMID: 17512403
- Weaver KE, Ehli EA, Nelson JS, Patel S. Antisense RNA regulation by stable complex formation in the Enterococcus faecalis plasmid pAD1 par addiction system. J Bacteriol 2004; 186:6400 - 8; http://dx.doi.org/10.1128/JB.186.19.6400-6408.2004; PMID: 15375120
- Ramirez-Peña E, Treviño J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol 2010; 78:1332 - 47; http://dx.doi.org/10.1111/j.1365-2958.2010.07427.x; PMID: 21143309
- Robertson HD. Escherichia coli ribonuclease III cleavage sites. Cell 1982; 30:669 - 72; http://dx.doi.org/10.1016/0092-8674(82)90270-7; PMID: 6754088
- de Smit MH, van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol 2003; 331:737 - 43; http://dx.doi.org/10.1016/S0022-2836(03)00809-X; PMID: 12909006
- Göbl C, Kosol S, Stockner T, Rückert HM, Zangger K. Solution structure and membrane binding of the toxin fst of the par addiction module. Biochemistry 2010; 49:6567 - 75; http://dx.doi.org/10.1021/bi1005128; PMID: 20677831
- Patel S, Weaver KE. Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis.. J Bacteriol 2006; 188:5374 - 84; http://dx.doi.org/10.1128/JB.00513-06; PMID: 16855226
- Weaver KE, Weaver DM, Wells CL, Waters CM, Gardner ME, Ehli EA. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J Bacteriol 2003; 185:2169 - 77; http://dx.doi.org/10.1128/JB.185.7.2169-2177.2003; PMID: 12644486
- Wu LJ, Errington J. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol 2012; 10:8 - 12; PMID: 22020262
- Pinho MG, Errington J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol 2003; 50:871 - 81; http://dx.doi.org/10.1046/j.1365-2958.2003.03719.x; PMID: 14617148
- Morlot C, Zapun A, Dideberg O, Vernet T. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol Microbiol 2003; 50:845 - 55; http://dx.doi.org/10.1046/j.1365-2958.2003.03767.x; PMID: 14617146
- Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A 1986; 83:3116 - 20; http://dx.doi.org/10.1073/pnas.83.10.3116; PMID: 3517851
- Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 2001; 276:1772 - 9; PMID: 11038353
- Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 2008; 72:579 - 89; http://dx.doi.org/10.1128/MMBR.00025-08; PMID: 19052321
- Palmer KL, Carniol K, Manson JM, Heiman D, Shea T, Young S, et al. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J Bacteriol 2010; 192:2469 - 70; http://dx.doi.org/10.1128/JB.00153-10; PMID: 20207762
- Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol 2002; 45:333 - 49; http://dx.doi.org/10.1046/j.1365-2958.2002.03042.x; PMID: 12123448
- Gerdes K, Wagner EGH. RNA antitoxins. Curr Opin Microbiol 2007; 10:117 - 24; http://dx.doi.org/10.1016/j.mib.2007.03.003; PMID: 17376733