Abstract
DEAD-box RNA helicases are present in almost all living organisms and participate in various processes of RNA metabolism. Bacterial proteins of this large family were shown to be required for translation initiation, ribosome biogenesis and RNA decay. The latter is primordial for rapid adaptation to changing environmental conditions. In particular, the RhlB RNA helicase from E. coli was shown to assist the bacterial degradosome machinery. Recently, the CshA DEAD-box proteins from Bacillus subtilis and Staphylococcus aureus were shown to interact with proteins that are believed to form the degradosome. S. aureus can cause life-threatening disease, with particular concern focusing on biofilm formation on catheters and prosthetic devices, since in this form the bacteria are almost impossible to eradicate both by the immune system and antibiotic treatment. This persistent state relies on the expression of surface encoded proteins that allow attachment to various surfaces, and contrasts with the dispersal mode of growth that relies on the secretion of proteins such as hemolysins and proteases. The switch between these two states is mainly mediated by the Staphylococcal cell density sensing system encoded by agr. We show that inactivation of the cshA DEAD-box gene results in dysregulation of biofilm formation and hemolysis through modulation of agr mRNA stability. Importantly, inactivation of the agrA gene in the cshA mutant background reverses the defect, indicating that cshA is genetically upstream of agr and that a delicate balance of agr mRNA abundance mediated through stability control by CshA is critical for proper expression of virulence factors.
Introduction
Tightly regulated RNA metabolism is important for cells to adapt to changing environments and to optimally utilize available nutrient resources. DEAD-box RNA helicases play crucial roles in almost all living organisms within the complexed interplay of RNA synthesis, RNA folding, RNA-RNA interactions, RNA localization, and, not least, RNA degradation. These RNA helicases are characterized by 12 conserved sequence motifs that are involved in ATP binding, RNA binding and intramolecular interactions.Citation1 These enzymes are able to locally unwind double stranded RNA in an ATP-dependent manner or they can clamp protein complexes to a substrate RNA under conditions in which ATP hydrolysis or Pi release are delayed.Citation1-Citation3
The model organisms, Escherichia coli and Bacillus subtilis, encode 5 and 4 DEAD-box proteins, respectively. Among the first DEAD-box proteins identified, the E. coli CsdA (previously called DeaD), SrmB, and DbpA were shown to be involved in ribosome biogenesis.Citation4-Citation6 The E. coli DbpA and the B. subtilis YxiN (DbpA) proteins are specifically stimulated by hairpin 92 of the 23S rRNA, and both interact with this substrate via their C-terminus.Citation7-Citation10 CsdA was also found to be involved in translation initiation.Citation11 The E. coli RhlE protein was proposed to be involved in ribosome biogenesis and interacts genetically with other DEAD-box proteins.Citation12 The E. coli RhlB protein was found to be part of the degradosome, which is composed of the scaffolding protein RNase E, PNPase and enolase.Citation13 The ATPase activity of recombinant RhlB protein is stimulated by RNase E.Citation14 An attractive hypothesis for the role of the RNA helicase RhlB within the degradosome would be the local unwinding of secondary structures that are inhibitory for RNA degradation by the PNPase.Citation13,Citation15 Additionally, CsdA was also found to be associated with the degradosome under cold shock conditions.Citation16
Several bacterial DEAD-box proteins were identified using a variety of screens, although the molecular basis for these phenotypes remains unclear. For example, DEAD-box proteins are required for phenolic acid metabolism, bacterial aggregation, oxidative stress response, cold adaption, and for growth in the absence of prey in the case of obligatory bacteriovorous bacteria.Citation17-Citation22 In Staphylococcus aureus, the DEAD-box protein CshA, was identified in a screen for mini-Mu insertions that reduced biofilm formation.Citation23 Furthermore, CshA and its homolog from B. subtilis were recently reported to interact with proteins present within the Gram-positive degradosome.Citation24,Citation25
S. aureus is an opportunistic pathogen carried asymptomatically by approximately one third of the human population. It can cause infections ranging from furuncles to severe osteomyelitis and endocarditis.Citation26 These very different clinical pathologies reflect the versatility of this bacterium to adapt to changing environments and its ability to switch from a harmless sessile state to a very aggressive and dispersive growth mode.Citation27 Importantly, S. aureus can cause highly persistent infections that are difficult or even impossible to eradicate and it is generally assumed that these are due to intracellular survival,Citation28 the formation of small colony variants,Citation29 or the presence of biofilms.Citation30 Biofilm formation can formally be divided into several stages.Citation31,Citation32 After primary adhesion via surface components, bacteria secrete proteins and exopolysaccharides to form an extracellular matrix, forming mushroom-like structures that eventually detach to seed new surfaces. This detachment is due to the presence of secreted proteases and surfactant peptides.Citation32
S. aureus uses a quorum-sensing system, agr, to regulate the switch from adhesive to dispersal behavior.Citation27 The agr operon encodes an auto-inducing peptide (AgrD) secreted by AgrB. At high AgrD concentrations, the peptide binds to the two-component system AgrC/AgrA. The AgrA protein then stimulates transcription of the agr operon resulting in a positive feedback loop and promotes synthesis of RNAIII, a regulatory RNA that positively and negatively regulates a variety of genes involved in virulence.Citation33 Several reports describe a function of the agr regulatory system in biofilm formation,Citation34 although this is markedly strain dependent.Citation35
Here we report that the CshA DEAD-box protein from S. aureus controls the stability of the agr mRNA, and thereby, virulence factor gene expression.
Results
CshA displays RNA-dependent ATPase activity
The S. aureus genome encodes two DEAD-box RNA helicases, SA1885 and SA1387, which are homologous to the B. subtilis CshA (YdbR) and CshB (YqfR), respectively (Fig. S1). The core sequence of the S. aureus CshA contains all the sequence motifs characteristic of DEAD-box proteins.Citation1 In addition, it possesses a highly charged (39+, 26-) region of 170 amino acids C-terminal of motif VI it that could be required for the interaction with RNA or other protein partners (Fig. S1).
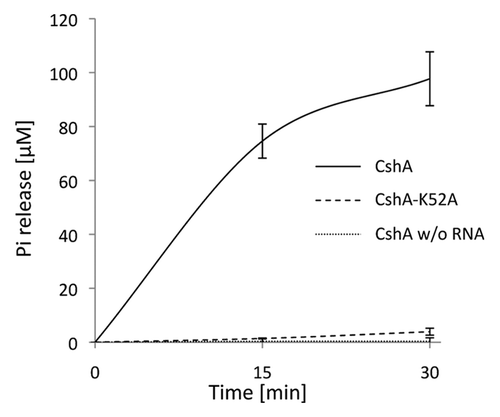
The B. subtilis CshA protein was shown to possess RNA dependent ATPase and dsRNA unwinding activity.Citation36 To assess the enzymatic activity of the staphylococcal CshA, we purified recombinant His6-tagged protein from E. coli and tested for ATPase activity.Citation37 CshA showed a clear RNA-dependant ATPase activity, typical of members of the DEAD-box family (). Michaelis-Menten kinetic analysis was performed using variable ATP concentrations under conditions of saturating RNA. The measured KM (ATP) value for CshA was 3.7 ± 0.47 mM with a Kcat value of 140 min−1. The ATPase activity of CshA carrying a mutation in motif I (Walker A motif; CshA-K52A; Fig. S1), that prevents ATP hydrolysis,Citation38,Citation39 was indistinguishable from that seen with buffer alone ().
The cshA mutant exhibits a cold-sensitive growth phenotype
Mutations in many bacterial RNA helicase genes cause a cold sensitive growth phenotype. We therefore tested growth of the cshA::mini-Mu mutant strains at different temperatures. Results demonstrated a cold-sensitive phenotype, with partial growth inhibition at 30°C and complete inhibition at room temperature (). To confirm that this phenotype was due to the mutation, we complemented the strain with the cshA gene under the control of a xylose-inducible promoter in the presence of 1% Xylose.Citation23 This restored growth to almost wild type at all temperatures tested.
Figure 2. Inactivation of the cshA gene results in a cold-sensitive growth phenotype. The parental wild type strain S30, the cshA::mini-Mu insertion, the mutant complemented with wild type cshA and a Walker A motif mutant (K52A) under the control of a xylose-inducible promoter were spotted in serial dilutions (indicated underneath) on rich medium containing 1% xylose and chloramphenicol (15 μg/ml). All were incubated at 42°C, 37°C, 30°C, and room temperature for 24 h, 24 h, 48 h, and 4 d, respectively. Plasmids expressing GFP instead of CshA were used as controls
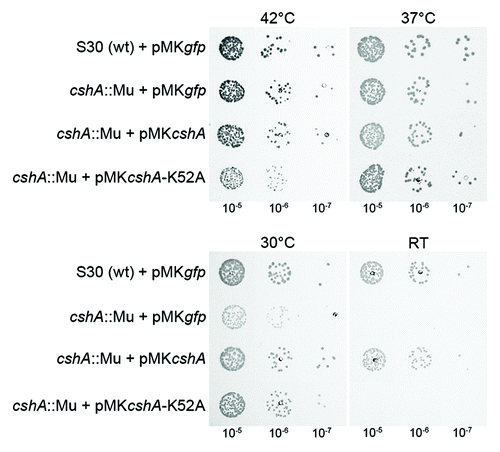
To ascertain that it is the enzymatic activity of the DEAD-box RNA helicase, and not a structural function within a complex, that was responsible for the observed phenotypes, we complemented the cshA::mini-Mu mutation with the K52A construct. Complementation by the mutant allele was not observed at room temperature (), although western blot analysis confirmed expression of the mutant protein (Fig. S2).
The cshA mutant is reduced in biofilm formation and shows increased hemolysis
The mini-Mu insertion in the cshA gene was originally isolated in a screen for biofilm-deficient mutants in the hyper-biofilm producing strain S30.Citation23 Biofilm formation in the mutant strain was reduced to approximately 30% of the wild type in microtiter plates or in polystyrene tubes, despite similar amounts of cells in the culture assay (). Complementation with a wild type cshA gene restored biofilm formation, whereas complementation with the cshA-K52A mutant did not, showing that enzymatic activity is needed for biofilm formation ().
Figure 3. CshA is required for biofilm formation and reduces hemolysis. (A) Biofilm formation was analyzed by the crystal violet (CV) assay in polystyrene tubes after growth for 6 h in TSB medium with equal amounts of inoculated cells. The amount of biofilm was measured after solubilization of the CV in ethanol and measuring the absorbance at 570 nm. The error bars (standard deviation) are from 3 independent experiments. The differences between S30 + pMKgfp vs cshA::Mu + pMKgfp and cshA::Mu pMKgfp vs cshA::Mu pMKcshA were significant (p = 0.001, p = 0.014, respectively). (B) S30 wild type strain and its cshA mutant derivative were spotted onto rabbit and horse blood agar plates. Shown is one representative result from 5 independent experiments. (C) Inactivation of agrA in the cshA mutant restores biofilm formation and reduces hemolysis.
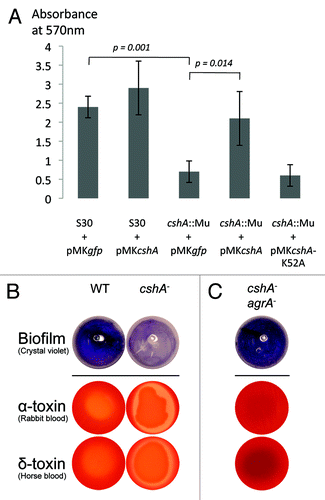
To further characterize the cshA::mini-Mu mutant, hemolysis was examined by spotting overnight cultures onto blood agar plates, which were incubated overnight at 37°C and then transferred to 4°C for a further two days to visualize hemolysis. The S30 cshA mutant showed drastically increased hemolysis on rabbit and horse blood plates, indicating an increase in α and delta toxin production, respectively (). Western blot analysis from exponential cultures confirmed the increased hemolysin synthesis (Fig. S4).
The agr mRNA is stabilized in the cshA mutant
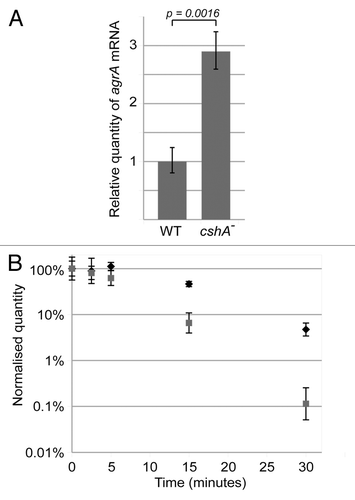
The reduction in biofilm formation and increase in hemolysis would be consistent with an upregulation of the agr mRNA and its RNAIII target.Citation40,Citation41 To test if this was indeed the case, we measured the steady-state levels of agrA mRNA by qRT-PCR in exponential phase cultures. The agrA mRNA was increased 2.9 fold in the cshA::mini-Mu mutant strain relative to the wild type ().
Figure 4. The agrA mRNA level is increased and stabilized in the cshA mutant strain. (A) RNA was isolated from exponentially growing cultures (approx. OD600 = 0.4) of the S30 wildtype and the cshA mutant. qRT-PCR was performed to determine the level of agr mRNA, using the HU mRNA as an internal reference. An unpaired T-test was performed to show that the difference in agr levels was significant (p = 0.0016). Error bars show the standard deviation. (B) Cultures of S30 (gray squares) and the cshA mutant (black diamonds) were rifampicin treated to block de novo RNA synthesis. Samples were taken for RNA isolation at 0, 2.5, 5, 15 and 30 min after treatment, and qRT-PCR was performed using primers and probe specific for agrA, and using HU mRNA as an internal reference. The quantity of agr, relative to HU, was normalized to 100% at time zero, and plotted in the graph. Error bars represent the 99% confidence level. The figure shows a single experiment out of three biological replicates.
In the light of the role of the E. coli RhlB DEAD-box protein in the degradosome and the bacterial two-hybrid interactions of the B. subtilis and S. aureus CshA proteins with components of the putative Gram-positive degradosome, the increase of the agrA mRNA might be caused by an mRNA degradation defect in the cshA mutant strain. To measure the stability of the agr mRNA in the parental and mutant strains, RNA was extracted at different time points after blocking transcription with rifampicin and agrA, agrB, and agrC mRNA levels were quantified by qRT-PCR using HU mRNA as an internal reference. The data show a significant stabilization of the agr mRNA in the cshA::mini-Mu mutant strain compared with its parental counterpart (; Fig. S5). Furthermore, the fact that all three analyzed regions of the agrBDCA operon mRNA behaved similarly makes a fortuitous stabilization of a particular region of the mRNA very unlikely. Thus, the stabilization of agr mRNA correlates with the steady-state increase of agr mRNA and, as shown below, also increases the level of RNAIII (Fig. S6E).
Inactivation of AgrA suppresses the cshA biofilm and hemolysis phenotypes
To check whether the biofilm defect is mediated through agr or via another CshA-dependent pathway, we disrupted the agrA open reading frame in the parental wild type and the cshA mutant strains using the targetron method.Citation42 Both biofilm formation and hemolysis on blood agar plates in the cshA agrA double mutant reverted to wild type (; Fig. S4), with a slight increase in biofilm formation and a complete abrogation of hemolysis.
The observed phenotypes can be reproduced in a different clinical isolate
The clinical strain S30 was observed to be a hyperbiofilm producer.Citation23 Therefore, to confirm that the observations made were not specific to this strain, we repeated the experiments with a different clinical isolate, SA564,Citation43 in which we inactivated the cshA gene by a targetron insertionCitation42 and by gene replacement using the pyrFE/5-FOA counter selection method.Citation44
The analysis showed that growth, biofilm formation, and agr RNA data were very similar to the S30 strain (Fig. S6). Deletion of cshA caused a cold sensitive phenotype for growth and, although SA564 does not produce a lot of biofilm, the cshA mutation reproducibly reduced biofilm formation as measured by the crystal violet assay. Most importantly, agr mRNA steady-state levels were increased and mRNA decay was reduced in the cshA mutant compared with the parent. These results show that the CshA-mediated regulation of agr occurs in diverse genetic backgrounds. Since an increase in agr levels should result in increased RNAIII expression, we also tested steady-state levels and RNAIII degradation after rifampicin treatment in the wild type and the cshA mutant strains. Whereas, as expected, the steady-state levels were increased, the rifampicin experiment revealed a rather stable RNAIII that was not further stabilized in absence of CshA (Fig. S6).
Discussion
We have shown that S. aureus cshA mutants are cold sensitive, growing poorly already at 30°C, display decreased biofilm production, and show increased hemolysin production. Cold sensitivity is observed for many DEAD-box helicase mutants across many species, and could be explained by the general inability to resolve one or more secondary RNA structures thereby blocking the expression of essential genes. Correspondingly, cshA was identified by transposon mutagenesis in a screen for genes that are important for growth at low temperatures in the food poisoning bacterium Bacillus cereus.Citation45,Citation46 Interestingly, a cshA mutation in Bacillus subtilis was reported to grow normally at low temperatures in one report,Citation47 but to be cold sensitive in another,Citation36 a discrepancy that may be strain dependent and needs further analysis.
We have demonstrated an increased stability of agr mRNA in the cshA mutant, which would account for the higher steady-state levels of the mRNA. This, in turn, induces synthesis of the regulatory RNAIII, an event that explains the increase of hemolysis and reduction of biofilm formation.Citation40,Citation41,Citation48
Biofilm formation and hemolysin production are directly, but in an opposing manner, influenced by a specific set of genes and conditions.Citation40,Citation49,Citation50 Since growth of the cshA mutant was almost like wild type at 37°C under conditions in which the amount of bacterial cells was carefully controlled, decreased biofilm was not the consequence of a general growth defect. Moreover, the increased hemolysin production in the mutant is in perfect agreement with the increased agr/RNAIII-levels.
To confirm that the observed hemolysis and biofilm phenotypes were indeed a result of changes in agr levels, and not due to another unknown effect of the cshA mutation, we constructed a cshA agrA double mutant that eliminated agr quorum sensing. In the double mutant we observed a higher level of biofilm formation compared with wild type and a complete abolishment of hemolysis. We propose that the complete block in agr function caused by the agrA mutation is the reason why the phenotypes of the agrA mutants appear to be more drastic than a simple reversion to wild type. Thus, the absence of the CshA DEAD-box protein affects the regulation of the central agr regulatory system and places cshA genetically upstream of agr. The observed phenotypes are in line with epidemiological observations that bacteria from persistent infections are often deficient in the agr regulon.Citation34,Citation51-Citation55 Nevertheless, an additional effect of CshA on gene expression downstream of the agr system cannot be excluded.
A role of CshA in RNA decay is consistent with recent bacterial two-hybrid data demonstrating that the CshA proteins from B. subtilis and S. aureus interact with components of a proposed Gram-positive degradosome.Citation24,Citation25 Interestingly, RNase J1 and RNase J2 were also identified with CshA in the screen for biofilm-deficient mutants.Citation23 The biochemical analysis of CshA from S. aureus showed low ATPase activity compared with other DEAD-box proteins.Citation56 This was approximately 1.8 fold higher in the presence of RNase J2 (data not shown), although a direct interaction between these proteins was not found in the bacterial two-hybrid system.Citation25 This stimulation is much weaker than the one observed for RhlB by RNase E.Citation14 At present the precise role of CshA in RNA degradation is not known but several hypotheses can be formulated. The DEAD-box protein could present a sequence specific interaction with target RNAs and thereby target RNases. However, so far, the only DEAD-box protein with clear sequence specificity is DbpA, involved in ribosome biogenesis. A more likely function could be in assisting the RNA turnover machinery to overcome inhibitory secondary structures. It was shown for the E. coli RhlB that this RNA helicase is required for RNAs with secondary structures to allow the activity of the PNPase to proceed. Future experiments will be necessary to assess the function of the DEAD-box protein in presence of one or more components of the putative degradosome.
Finally, regulation of the quorum sensing system by RNA turnover may be a more general phenomenon. Indeed, in order to change quickly from high-level agr expression to low level of quorum sensing readout, it will be necessary to rapidly turn down the system (). Otherwise a bacterial cell that is released from a dense population of Staphylococcal cells, would not sufficient rapidly be able to turn on surface protein expression, necessary for adhesion and immune escape. Diluting out the mRNA by cell division or degradation with the bulk mRNA pool are probably not sufficient to obtain a rapid adaption to a new situation. Further analyses in Staphylococcus and other bacteria may reveal other similar situations, where RNA turnover is primordial to rapid adaptation.
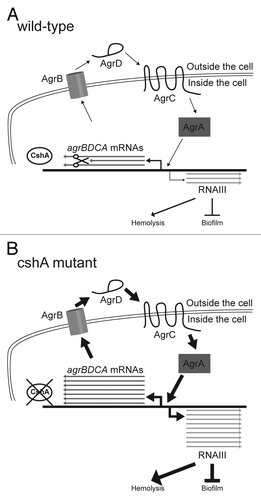
Figure 5. Model of CshA in the degradation of the agr mRNA. (A) In the wild type, agrBDCA mRNAs are produced. However, in the presence of CshA we hypothesize that the degradosome is able to degrade a significant portion of them. Thus the quorum sensing system is working correctly and only small amounts of RNAIII is produced leading to low stimulation of hemolysis and normal biofilm formation. (B) In absence of CshA, we propose that the degradosome is unable to degrade the agrBDCA mRNAs correctly, leading to a much higher level of Agr proteins, resulting in elevated RNAIII levels. The RNAIII, in turn, strongly stimulates production of hemolysins and extracellular proteases, and inhibits the production of biofilm components.
In conclusion, our data show that the CshA DEAD-box protein is an important player in the regulation of S. aureus virulence factors through its involvement in mRNA turnover, in particular the mRNA of the central regulator agr. Whereas a DEAH-box RNA helicase was found involved in global gene regulation in Borrelia burgdorferiCitation57 and a DEAD-box protein of the Dhh1 subfamily in regulation of virulence associated genes in the pathogenic fungus Cryptococcus neoformans Citation57, it is to the best of our knowledge the first time that a DEAD-box RNA helicase is implicated in the regulation of virulence factor expression in a pathogenic bacterium and that a functional CshA protein is important for the regulatory role of the agr quorum sensing system in S. aureus.
Material and Methods
Strains and plasmids
Bacteria were grown under standard laboratory conditions.Citation23,Citation58 For the inactivation of the agrA gene, a targetron construct for the integration of a type II intron in anti-sense orientation was generated in a pNL9162 plasmid,Citation42 and a SphI-NarI fragment containing the intron and the intron encoded protein (IEP) was recloned into the SmaI site of pMK4Citation59 to avoid incompatibility problems between pNL9162 and the naturally resident pT181 plasmid in S30. After transformation of the parental and the S30 cshA::mini-Mu strain with this pMK4-agrA-targetron plasmid, colonies were screened by PCR for disruption of agrA by integration of the intron after coordinate 442 of the agrA open reading frame. The strain PR01ΔcshA was constructed using the pyrFE/5-FOA counter-selection system.Citation44 For complementation, the cshA gene was cloned under control of a xylose inducible promoter in plasmid pMK4Citation23 into the KpnI and PstI sites using PCR amplified DNA from strain S30, resulting in pMK4cshA. For the construction of the K52A mutant, a fusion-PCR product was made using the and the same external oligonuclotides resulting in pMK4cshA-K52A. Strains and plasmids are listed in and . Oligonucleotides are listed in supporting . For overexpression in E. coli, the CshA open reading frame was PCR amplified and cloned into the NdeI and BamHI sites of pET22b (Novagen). For the recombinant K52A mutant gene, the same K52A-mutant oligonucleotides were used.
Table 1. Strains
Table 2. Plasmids
Standard molecular biology methods for plasmid and strain constructions were employed according to either the manufacturer’s instructions or Sambrook and Russell.Citation58
Purification of proteins and ATPase activity
Overexpression and purification of CshA and CshA-K52A were performed as follows: overnight culture of E. coli Rosetta (DE3) carrying pET22b-CshA was diluted in 200 mL LB with 100 µg/mL ampicillin and grown at 30°C. Expression of the protein was induced with 0.5 mM IPTG at OD600 0.5 and incubated for 2h. Purification of proteins was adapted from Tanner et al.Citation37 To check the purity of the protein, an aliquot of each eluate was loaded on a 10% SDS-PAGE. ATPase activity was tested at different pH (6.5, 7.5, 8), in different buffers (potassium acetate HEPES, KCl TRIS-HCl, potassium actate TRIS-HCl), different potassium acetate concentrations (40, 50, 60 and 80 mM), and different magnesium acetate concentrations (1, 2, 3, and 4 mM). Optimal reaction conditions for CshA activity were determined to be 50 mM potassium acetate, 20 mM HEPES (pH 7.5), 2 mM magnesium acetate at 37°C. ATPase activity in the presence of 1 mM ATP was measured using the Malachite green method for detection of released Pi and monitored at 630 nm.Citation37 Michaelis-Menten kinetics of the ATPase reaction were performed using 70 nM of CshA protein, in presence of saturating E. coli rRNA (1 µg/µL) and varying ATP concentration from 0 to 4 mM. KM and Kcat were calculated using Kaleidagraph software (Synergy). The standard deviations were derived from the curve fits based on three independent sets of experiments.
Biofilm formation
Optical densities (OD) of bacteria from over night culture grown at 37°C with agitation were measured at 600nm. Bacterial suspensions were diluted to an OD of 0.1 into 1 ml fresh TSB medium and inoculated in polystyrene tubes (BD Biosciences 352057). After incubation for 6 h at 37°C without agitation, biofilms attached to the surface were stained for 10 min with a solution of 1% (wt/vol) Crystal Violet (CV) 2% (vol/vol) ethanol, rinsed carefully with water, and air-dried. Residual biofilm-associated CV dye was dissolved with 400 μl ethanol, diluted with 600 μl H2O and transferred to a new tube for absorbance measurement at 570nm.
Hemolysin assay
Mueller-Hinton medium with 10 g/l agar was autoclaved and cooled to 43°C, whereupon 50 ml/l 40% xylose and 70 ml/l blood (preheated to 43°C) was added, and the plates were immediately poured. De-fibrinated rabbit and horse blood was obtained from TSC Biosciences (Buckingham, UK). Spots of 10 μl over night culture were deposited on the plates and allowed to dry, incubated for about 20 h at 37°C, and two days at 5°C before scoring the extent of the hemolysis zone. At least five independent assays were performed, with mutants and wild type always on the same plate.
For Western Blot analysis, overnight cultures were diluted in MH medium to 1/100. Ten ml of each culture were collected after 4h (exponential phase ; 0D = 0.6) of growth at 37°C under shaking conditions. Samples were centrifuged at 10000 rpm and supernatants were incubated in the presence of Trichloroacetic acid (12% final) at 4°C overnight. Precipitates were centrifuged at 14000 rpm, washed three times in acetone, resuspended in 50 µl Laemmli buffer and migrated on a 10% PAGE. Western blot analysis was performed with anti-Staphylococcus α hemolysin antibodies (abcam, ab50536) diluted to 1/20000, according to the manufacturer’s instructions.
Choice of reference for RNA quantification
Since it cannot be excluded that CshA also participates in ribosome biogenesis and since the 16S rRNA has extensive secondary structure, which might possibly be interacting with an RNA helicase such as CshA, we chose a highly stable mRNA, HU, which has been used as a reference in other studies.Citation60 To evaluate whether HU mRNA remains mostly unaffected by the cshA mutation, the decay-rate of HU mRNA was compared with ten other mRNAs (asp23, deoD, dapA, dapB, asd, lytM, ilvC, sspA, purA) by qRT-PCR in a manner similar to the agrBDCA operon. Out of the ten, which were chosen by using primers and TagMan-probes that were already available from other projects, six (asp23, dapB, deoD, dapA, asd, lytM) exhibited similar decay characteristics as HU mRNA, i.e., no change between wild type and cshA mutant when compared with the HU mRNA (data not shown). To further validate the use of HU as reference mRNA, we measured the amount of HU mRNA in the total RNA pool and found it to be relatively stable and unaffected by the cshA mutation (Fig S3).
RNA stability assay
S. aureus strains from 2 ml overnight cultures grown in MHB were diluted 1:200 into 20 ml of fresh medium and were incubated at 37°C under agitation. At an OD600 = 0.4 (in some experiments after 4h), Rifampicin (200 µg/ml) was added to the culture and 1 ml was taken at different time points: 0, 2.5, 5, 15 and 30 min. Each sample was immediately centrifuged for 20 sec at 8000 g and then the pellet was rapidly fixed in 500 µl of acetone:ethanol (1:1) and stored at -80°C. Total RNA was extracted as described below. For SA564, we performed the assays using 400 µg/ml antibiotic. In SA564 wild type the agr mRNA levels were, under these conditions, down to basal level after only 15 min, and therefore the time points taken were at 0, 2.5, 5, 10 and 15 min.
Total RNA extraction
Bacterial samples were harvested and washed in TE buffer (10 mM TRIS-HCl, 1 mM EDTA [pH 8.0]). Samples were lysed in TE containing lysostaphin (250 µg/ml) for 10 min at 37°C. RNA was extracted using the RNeasy mini Kit (Qiagen, 74104) and QIAshredder columns (Qiagen, 79654) following the manufacturer's instructions. DNA was removed from the RNA preparations by treatment with DNase I (Qiagen, 79254). Purified RNA samples were analyzed by using the RNA NanoLab chip kit (Agilent) on the 2100 Bioanalyser instrument (Agilent). At least two independent cultures and RNA extractions were performed for each strain.
Real-time RT-PCR
mRNA levels were determined by quantitative RT-PCR using the Brilliant II QRT-PCR Master Mix Kit, 1-Step (Stratagene). Primers and probes (Supporting ) were designed using the PrimerExpress software 3.0 (Applied Biosystems) and obtained from Sigma, Applied Biosystems or Eurogentec. Reactions were performed using 1.25 ng of total RNA in a final volume of 10 µl, with the primers and probes (Supporting ) at concentrations of 0.2 µM and 0.1 µM, respectively. RT-PCR mixtures were incubated for 30 min at 50°C, followed by incubation for 10 min at 95°C and then 40 cycles of 15 sec at 95°C and 1 min at 60°C in a StepOne Plus instrument (Applied Biosystems). The mRNA levels of target genes extracted from the different strains were normalized relative to their HU mRNA levels (see above). To minimize experimental variations, samples taken at different times from the same culture were treated simultaneously (RNA extraction and qRT-PCR analysis).
Additional material
Download Zip (4.7 MB)Acknowledgments
We are grateful to Sanda Rocak for initial experiments, Josette Banroques and N. Kyle Tanner for initial help with the purification of CshA, William Kelley for discussions and Patrick Viollier, Urs Jenal and Joseph Curran for helpful comments on the manuscript. This work was supported by grants of the Swiss National Science Foundation to P.L., J.S. and P.F., by the Roche Research Foundation (P.L.), the E. and L. Schmidheiny Foundation (P.L.), and the Canton of Geneva.
Author Contributions
Conceived and designed the experiments: P.R., P.F., J.S., P.L. Designed reference gene: P.F., J.S. Performed the experiments: S.O., P.R., A.C., J.P.D., E.B., C.G., M.G. Analyzed the data: P.R., J.P.D., P.F., J.S., P.L. Wrote the paper: P.R., P.L.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 2011; 12:505 - 16; http://dx.doi.org/10.1038/nrm3154; PMID: 21779027
- Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A 2008; 105:20209 - 14; http://dx.doi.org/10.1073/pnas.0811115106; PMID: 19088201
- Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res 2006; 34:4181 - 8; http://dx.doi.org/10.1093/nar/gkl410; PMID: 16935886
- Nishi K, Morel-Deville F, Hershey JWB, Leighton T, Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature 1988; 336:496 - 8; http://dx.doi.org/10.1038/336496a0; PMID: 2461520
- Toone WM, Rudd KE, Friesen JD. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol 1991; 173:3291 - 302; PMID: 2045359
- Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res 2009; 37:6503 - 14; http://dx.doi.org/10.1093/nar/gkp711; PMID: 19734347
- Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J 1993; 12:3619 - 26; PMID: 8253085
- Tsu CA, Uhlenbeck OC. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry 1998; 37:16989 - 96; http://dx.doi.org/10.1021/bi981837y; PMID: 9836593
- Kossen K, Uhlenbeck OC. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res 1999; 27:3811 - 20; http://dx.doi.org/10.1093/nar/27.19.3811; PMID: 10481020
- Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA 2006; 12:959 - 67; http://dx.doi.org/10.1261/rna.5906; PMID: 16611943
- Moll I, Grill S, Gründling A, Bläsi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol 2002; 44:1387 - 96; http://dx.doi.org/10.1046/j.1365-2958.2002.02971.x; PMID: 12068815
- Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA 2008; 14:381 - 9; http://dx.doi.org/10.1261/rna.800308; PMID: 18083833
- Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 1996; 381:169 - 72; http://dx.doi.org/10.1038/381169a0; PMID: 8610017
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, et al. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev 1998; 12:2770 - 81; http://dx.doi.org/10.1101/gad.12.17.2770; PMID: 9732274
- Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev 1999; 13:2594 - 603; http://dx.doi.org/10.1101/gad.13.19.2594; PMID: 10521403
- Prud’homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol Microbiol 2004; 54:1409 - 21; http://dx.doi.org/10.1111/j.1365-2958.2004.04360.x; PMID: 15554978
- Gury J, Barthelmebs L, Cavin JF. Random transposon mutagenesis of Lactobacillus plantarum by using the pGh9:IS S1 vector to clone genes involved in the regulation of phenolic acid metabolism. Arch Microbiol 2004; 182:337 - 45; http://dx.doi.org/10.1007/s00203-004-0705-1; PMID: 15375644
- Pandiani F, Chamot S, Brillard J, Carlin F, Nguyen-the C, Broussolle V. Role of the five RNA helicases in the adaptive response of Bacillus cereus ATCC 14579 cells to temperature, pH, and oxidative stresses. Appl Environ Microbiol 2011; 77:5604 - 9; http://dx.doi.org/10.1128/AEM.02974-10; PMID: 21705526
- Roos S, Lindgren S, Jonsson H. Autoaggregation of Lactobacillus reuteri is mediated by a putative DEAD-box helicase. Mol Microbiol 1999; 32:427 - 36; http://dx.doi.org/10.1046/j.1365-2958.1999.01363.x; PMID: 10231497
- Briolat V, Reysset G. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J Bacteriol 2002; 184:2333 - 43; http://dx.doi.org/10.1128/JB.184.9.2333-2343.2002; PMID: 11948145
- Markkula A, Mattila M, Lindström M, Korkeala H. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3°C. Environ Microbiol 2012; 14:2223 - 32; http://dx.doi.org/10.1111/j.1462-2920.2012.02761.x; PMID: 22564273
- Pearce BJ, Yin YB, Masure HR. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol 1993; 9:1037 - 50; http://dx.doi.org/10.1111/j.1365-2958.1993.tb01233.x; PMID: 7934910
- Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, et al. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus.. Infect Immun 2007; 75:1079 - 88; http://dx.doi.org/10.1128/IAI.01143-06; PMID: 17158901
- Lehnik-Habrink M, Pförtner H, Rempeters L, Pietack N, Herzberg C, Stülke J. The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol Microbiol 2010; 77:958 - 71; PMID: 20572937
- Roux CM, DeMuth JP, Dunman PM. Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J Bacteriol 2011; 193:5520 - 6; http://dx.doi.org/10.1128/JB.05485-11; PMID: 21764917
- Lowy FD. Staphylococcus aureus infections. N Engl J Med 1998; 339:520 - 32; http://dx.doi.org/10.1056/NEJM199808203390806; PMID: 9709046
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 2003; 48:1429 - 49; http://dx.doi.org/10.1046/j.1365-2958.2003.03526.x; PMID: 12791129
- Garzoni C, Kelley WL. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol 2009; 17:59 - 65; http://dx.doi.org/10.1016/j.tim.2008.11.005; PMID: 19208480
- von Eiff C. Staphylococcus aureus small colony variants: a challenge to microbiologists and clinicians. Int J Antimicrob Agents 2008; 31:507 - 10; http://dx.doi.org/10.1016/j.ijantimicag.2007.10.026; PMID: 18180148
- Goerke C, Wolz C. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int J Med Microbiol 2010; 300:520 - 5; http://dx.doi.org/10.1016/j.ijmm.2010.08.003; PMID: 20843740
- Götz F. Staphylococcus and biofilms. Mol Microbiol 2002; 43:1367 - 78; http://dx.doi.org/10.1046/j.1365-2958.2002.02827.x; PMID: 11952892
- Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol 2008; 322:207 - 28; http://dx.doi.org/10.1007/978-3-540-75418-3_10; PMID: 18453278
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 2007; 21:1353 - 66; http://dx.doi.org/10.1101/gad.423507; PMID: 17545468
- Vuong C, Götz F, Otto M. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect Immun 2000; 68:1048 - 53; http://dx.doi.org/10.1128/IAI.68.3.1048-1053.2000; PMID: 10678906
- O’Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, et al. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol 2007; 45:1379 - 88; http://dx.doi.org/10.1128/JCM.02280-06; PMID: 17329452
- Ando Y, Nakamura K. Bacillus subtilis DEAD protein YdbR possesses ATPase, RNA binding, and RNA unwinding activities. Biosci Biotechnol Biochem 2006; 70:1606 - 15; http://dx.doi.org/10.1271/bbb.50678; PMID: 16861794
- Tanner NK, Cordin O, Banroques J, Doère M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell 2003; 11:127 - 38; http://dx.doi.org/10.1016/S1097-2765(03)00006-6; PMID: 12535527
- Rozen F, Pelletier J, Trachsel H, Sonenberg N. A lysine substitution in the ATP-binding site of eucaryotic initiation factor 4A abrogates nucleotide-binding activity. Mol Cell Biol 1989; 9:4061 - 3; PMID: 2506440
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 2006; 125:287 - 300; http://dx.doi.org/10.1016/j.cell.2006.01.054; PMID: 16630817
- Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 1993; 12:3967 - 75; PMID: 7691599
- Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J 1990; 9:1391 - 9; PMID: 2328718
- Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 2006; 12:1271 - 81; http://dx.doi.org/10.1261/rna.68706; PMID: 16741231
- Somerville GA, Beres SB, Fitzgerald JR, DeLeo FR, Cole RL, Hoff JS, et al. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J Bacteriol 2002; 184:1430 - 7; http://dx.doi.org/10.1128/JB.184.5.1430-1437.2002; PMID: 11844774
- Redder P, Linder P. New range of vectors with a stringent 5-fluoroorotic acid-based counterselection system for generating mutants by allelic replacement in Staphylococcus aureus. Appl Environ Microbiol 2012; 78:3846 - 54; http://dx.doi.org/10.1128/AEM.00202-12; PMID: 22447609
- Broussolle V, Pandiani F, Haddad N, Michaud C, Carlin F, Nguyen-the C, et al. Insertional mutagenesis reveals genes involved in Bacillus cereus ATCC 14579 growth at low temperature. FEMS Microbiol Lett 2010; 306:177 - 83; http://dx.doi.org/10.1111/j.1574-6968.2010.01953.x; PMID: 20370835
- Pandiani F, Brillard J, Bornard I, Michaud C, Chamot S, Nguyen-the C, et al. Differential involvement of the five RNA helicases in adaptation of Bacillus cereus ATCC 14579 to low growth temperatures. Appl Environ Microbiol 2010; 76:6692 - 7; http://dx.doi.org/10.1128/AEM.00782-10; PMID: 20709848
- Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J Bacteriol 2006; 188:240 - 8; http://dx.doi.org/10.1128/JB.188.1.240-248.2006; PMID: 16352840
- Kong KF, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol 2006; 296:133 - 9; http://dx.doi.org/10.1016/j.ijmm.2006.01.042; PMID: 16487744
- Cafiso V, Bertuccio T, Santagati M, Demelio V, Spina D, Nicoletti G, et al. agr-Genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol Med Microbiol 2007; 51:220 - 7; http://dx.doi.org/10.1111/j.1574-695X.2007.00298.x; PMID: 17854479
- Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 2008; 4:e1000052; http://dx.doi.org/10.1371/journal.ppat.1000052; PMID: 18437240
- Goerke C, Campana S, Bayer MG, Döring G, Botzenhart K, Wolz C. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun 2000; 68:1304 - 11; http://dx.doi.org/10.1128/IAI.68.3.1304-1311.2000; PMID: 10678942
- Sakoulas G, Eliopoulos GM, Moellering RC Jr., Wennersten C, Venkataraman L, Novick RP, et al. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 2002; 46:1492 - 502; http://dx.doi.org/10.1128/AAC.46.5.1492-1502.2002; PMID: 11959587
- Fowler VG Jr., Sakoulas G, McIntyre LM, Meka VG, Arbeit RD, Cabell CH, et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 2004; 190:1140 - 9; http://dx.doi.org/10.1086/423145; PMID: 15319865
- Vuong C, Dürr M, Carmody AB, Peschel A, Klebanoff SJ, Otto M. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol 2004; 6:753 - 9; http://dx.doi.org/10.1111/j.1462-5822.2004.00401.x; PMID: 15236642
- Yarwood JM, Paquette KM, Tikh IB, Volper EM, Greenberg EP. Generation of virulence factor variants in Staphylococcus aureus biofilms. J Bacteriol 2007; 189:7961 - 7; http://dx.doi.org/10.1128/JB.00789-07; PMID: 17675387
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene 2006; 367:17 - 37; http://dx.doi.org/10.1016/j.gene.2005.10.019; PMID: 16337753
- Steimer L, Klostermeier D. RNA helicases in infection and disease. RNA Biol 2012; 9:751 - 71; http://dx.doi.org/10.4161/rna.20090; PMID: 22699555
- Sambrook J, Russell DR. Molecular cloning: a laboratory manual. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press, 2001.
- Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 1984; 29:21 - 6; http://dx.doi.org/10.1016/0378-1119(84)90161-6; PMID: 6092222
- Chien Y, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem 1999; 274:37169 - 76; http://dx.doi.org/10.1074/jbc.274.52.37169; PMID: 10601279
- Redder P, Linder P. New range of vectors with a stringent 5-fluoroorotic acid-based counterselection system for generating mutants by allelic replacement in Staphylococcus aureus. Appl Environ Microbiol 2012; 78:3846 - 54; http://dx.doi.org/10.1128/AEM.00202-12; PMID: 22447609
- Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 2004; 70:6076 - 85; http://dx.doi.org/10.1128/AEM.70.10.6076-6085.2004; PMID: 15466553