Abstract
Genuine La and La-related proteins group 7 (LARP7) bind to the non-coding RNAs transcribed by RNA polymerase III (RNAPIII), which end in UUU-3′OH. The La motif and RRM1 of these proteins (the La module) cooperate to bind the UUU-3′OH, protecting the RNA from degradation, while other domains may be important for RNA folding or other functions. Among the RNAPIII transcripts is ciliate telomerase RNA (TER). p65, a member of the LARP7 family, is an integral Tetrahymena thermophila telomerase holoenzyme protein required for TER biogenesis and telomerase RNP assembly. p65, together with TER and telomerase reverse transcriptase (TERT), form the Tetrahymena telomerase RNP catalytic core. p65 has an N-terminal domain followed by a La module and a C-terminal domain, which binds to the TER stem 4. We recently showed that the p65 C-terminal domain harbors a cryptic, atypical RRM, which uses a unique mode of single- and double-strand RNA binding and is required for telomerase RNP catalytic core assembly. This domain, which we named xRRM, appears to be present in and unique to genuine La and LARP7 proteins. Here we review the structure of the xRRM, discuss how this domain could recognize diverse substrates of La and LARP7 proteins and discuss the functional implications of the xRRM as an RNP chaperone.
Introduction
Non-coding RNAs transcribed by RNAPIII in eukaryotes terminate in a UUU-3′OH that is recognized by the ubiquitous La protein, which binds to the UUU-3′OH to prevent exonucleolytic degradation ().Citation1 Further processing of RNAs removes the UUU-3′OH and hence La is usually not part of the final RNA-protein complexes (RNPs) of these RNAs.Citation2,Citation3 The La protein is replaced in some non-coding RNAs, including ciliate telomerase RNA and cellular 7SK RNA, by the LARP7 class of proteins. LARP7 share the domain architecture of genuine La proteins but have evolved specific RNA binding and chaperoning functions, in addition to UUU-3′OH binding ().Citation2,Citation3
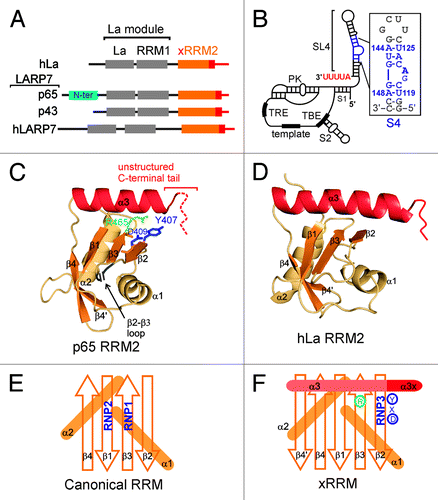
Figure 1. hLa and LARP7 proteins and Tetrahymena telomerase RNA. (A) Domain architecture of hLa, Tetrahymena p65, Euplotes p43 and human 7SK LARP7. Telomerase LARP7 p65 has a Tetrahymena specific N-terminal domain (in cyan) that is absent in other LARP7 proteins. (B) The secondary structure and conserved elements in Tetrahymena TER: S1 is stem 1, S2 is stem 2, SL4 is stem-loop 4, S4 is stem 4, TBE is template boundary element, PK is pseudoknot and TRE is template recognition element. The secondary structure and nucleotide sequence of the S4 construct used in this study is shown in the inset. The native nucleotides are in blue with numbering from full-length TER and the two (non-conserved) G-C base pairs at the bottom and terminal G(UUCG)C loop at the top are in gray. (C) The structure of p65 xRRM2 domain (PDB ID 4EYT). All the secondary structure elements are marked. The positions of the 46 residue β2-β3 loop and the unstructured C-terminal tail are marked. The RNP3 residues Y407 and D409 on β2, and conserved R465 on β3 are marked. (D) Structure of hLa RRM2 structure (PDB ID 1OWX). All the secondary structure elements are marked. The non-canonical helix α3 is in red. (E) Schematic representation of a canonical RRM with the position of RNP1 and RNP2 shown. (F) Schematic representation of xRRM with position of RNP3, conserved R465 and helix α3x shown. Residues of RNP3 in p65 xRRM2 (Y-X-D) shown.
In Tetrahymena, p65, which is a LARP7, is an integral protein of the telomerase holoenzyme. Telomerase is a unique reverse transcriptase that is responsible for maintenance of telomere repeat DNA in most eukaryotes.Citation4,Citation5 The telomerase holoenzyme is an RNP, which has an non-coding RNA component called telomerase RNA (TER), a specialized telomerase reverse transcriptase (TERT) and several species-specific accessory proteins.Citation5,Citation6 The accessory proteins are involved in regulation of holoenzyme biogenesis, assembly, localization and activity in vivo.Citation7 TER provides a template for telomere repeat synthesis by TERT and also acts as a scaffold on which the telomerase proteins assemble ().Citation5 A large part of our understanding of telomerase assembly, structure and function comes from the model organism Tetrahymena thermophila, a ciliated protozoan. Tetrahymena telomerase is the best characterized telomerase holoenzyme in terms of its subunit composition, assembly mechanism and activity.Citation5,Citation8,Citation9
Although in vitro only TER and TERT are required to reconstitute telomerase catalytic activity, in vivo the RNP catalytic core contains p65 in addition to TER and TERT.Citation10,Citation11 The p65 interacts with stem 4 (S4), UUU-3′OH and possibly stem 1 of TER, which are distinct from the TERT binding sites ().Citation10-Citation14 The interaction of p65 with TER is essential for catalytic core assembly in vivoCitation10,Citation11 and enhances assembly and activity in vitro.Citation10,Citation12,Citation15 An extensive network of biochemical interactions has been established between the three RNP catalytic core components, p65-TER-TERT, which contribute to different aspects of telomerase RNP assembly and function.Citation10,Citation11,Citation15,Citation16 Some mutations in TER and TERT that abrogate or decrease activity in telomerase assembled in vitro from TERT and TER are rescued when p65 is present, which further underscores the essential role of p65 in the biogenesis and assembly of the telomerase holoenzyme.Citation15,Citation17 The interaction of p65 with TER is discussed next in detail.
p65 is a telomerase catalytic core protein
Genetic depletion of p65 in Tetrahymena leads to decreased levels of both TER and TERT, indicating its critical role in Tetrahymena telomerase biogenesis and assembly.Citation8,Citation10,Citation11 p65 consists of four domains: N-terminal domain, La motif, RRM1 and C-terminal domain (). We recently showed that this C-terminal domain is a novel RRM, named xRRM.Citation13 The N-terminal domain is predicted to have a helical fold (data not shown); however, its precise role in p65 function is not clear. The La motif and RRM1 together constitute a La module, which is a conserved module in genuine La and La-related proteins ().Citation2,Citation3,Citation18 In all La family proteins, the La module has been implicated in initial recognition and binding to the UUU-3′OH sequence of various RNAPIII transcripts (and some intermediate RNA Polymerase II transcripts in yeast).Citation2,Citation3,Citation18 Hence, the p65 La module is implicated in recognition of TER UUU-3′OH, protecting it from exonuclease degradation. p65 C-terminal xRRM2 binds with high affinity and specificity to the TER S4, including the conserved GA bulge nucleotides ().Citation10,Citation13 The structure of the xRRM in complex with TER S4 RNA and other studies of this domain have shown that it is necessary and sufficient for inducing a conformational change in TER required for assembly of TERT with p65-TER to form the p65-TER-TERT ternary catalytic core.Citation10,Citation13,Citation14
The other known proteins belonging to the LARP7 family are p43 from ciliate Euplotes telomerase and the human 7SK RNP protein hLARP7 (). p43 is a telomerase assembly protein in Euplotes homologus to p65 in Tetrahymena.Citation19 hLARP7 binds the 3′ end of 7SK RNA and helps in its folding, stability and subsequent recognition of positive transcription elongation factor b (P-TEFb).Citation13,Citation20,Citation21 The 7SK RNP plays an essential role in regulation of transcription of all RNA polymerase II mRNA transcripts by sequestering P-TEFb in the RNP, thereby limiting the amount of active free P-TEFb in the cell.Citation22 P-TEFb is a cyclin-dependent kinase that plays an essential role in transcription regulation by RNA polymerase II.
p65 C-terminal domain contains an atypical, cryptic RRM similar to hLa RRM2
Recently, we determined the solution and crystal structures of the C-terminal domain of Tetrahymena telomerase p65 ().Citation13 This is the first structure of the C-terminal domain of any LARP7. The structure consists of an atypical RRM fold of βαββαβ’βα topology with a novel binding mode that we identified as a new class of RRM, the xRRM (see below). The RRM is one of the most abundant domains in proteins of the eukaryotic genome.Citation23,Citation24 A canonical RRM has a βαββαβ topology with two conserved sequence motifs: RNP1 on β3 [sequence (R/K)-G-(F/Y)-(G/A)-(F/Y)-(I/L/V)-X-(F/Y)] and RNP2 on β1 [sequence (I/L/V)-(F/Y)-(I/L/V)-X-N-L] ().Citation23,Citation24 The four β-strands form a β-sheet and two α-helices are packed on the “back side” of the β-sheet forming a compact fold. The conserved RNP1 and RNP2 residues located on the solvent exposed “front face” of the β-sheet generally contribute to specific recognition of single-stranded RNA.Citation23,Citation24 Usually, additional solvent exposed residues on the β-sheet and/or the loops are also involved in specific recognition of single-stranded RNA.Citation24In contrast, the p65 RRM2 has the atypical RRM features of hLa RRM2, including the presence of an additional β4’, absence of the canonical RNP1 and RNP2 sequences and presence of a non-canonical helix α3 lying across the β-sheet ().Citation13,Citation25 The p65 xRRM2 has two additional unusual features: an unusually long flexible loop (~45 residues) between the β2-β3 strands (β2-β3 loop) and an intrinsically disordered C-terminal tail (). The presence of the unusually long β2-β3 loop in the middle of the domain made it “cryptic” and explains why it was not predicted as an RRM fold.Citation13 The long β2-β3 loop appears to be unique to Tetrahymena telomerase p65, as it is not present in the homologous Euplotes telomerase protein p43.Citation19
Identification of xRRM: An RRM with extended α3 upon RNA binding
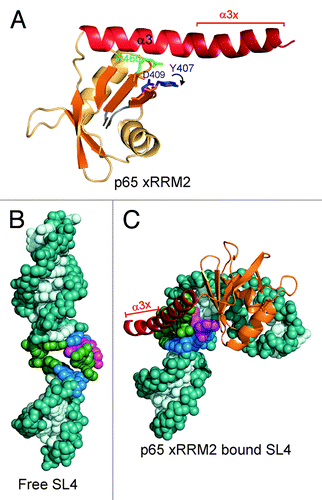
The crystal structure of p65 xRRM2 in complex with TER S4 provided the first structure of an RRM2 from any La or LARP7 protein in complex with RNA ().Citation13 This novel RRM interacts with both single- and double-stranded RNA using the β-sheet surface and the C-terminal tail, which forms a helical extension of α3 (α3x) that binds to the RNA major groove. We named this domain xRRM, for atypical RRM with extended α3 (xRRM2) (). Binding of xRRM2 to the conserved GA bulge and the major groove results in a large (105°) roll between the two conserved G-C base pairs flanking the GA bulge (). Comparison to the free TER stem-loop 4 (SL4) shows the dramatic conformational change, from a relatively straight RNA to the highly bent RNA in complex with p65 xRRM2.Citation13,Citation26 The structure provided insight into the structural basis for Tetrahymena telomerase catalytic core assembly.Citation13
Figure 2. The p65 xRRM2 and its interaction with RNA. (A) The structure of p65 xRRM2 domain in the RNA-bound state. All the secondary structure elements are the same as in . The C-terminal residues that form a helix upon binding to the double-stranded RNA are marked as α3x. The position of the key RNP3 residues Y407 and D409, and conserved R465 in the bound state are marked. A curved arrow depicts the movement of Y407 side-chain upon stacking on G121 of TER S4. (B and C) Conformational change in TER SL4 upon p65 xRRM2 binding. (B) Free structure of SL4 of Tetrahymena TER (based on PDB ID 2FEY).Citation26 (C) Model of TER SL4 bound to p65 xRRM2. The model is based on p65 xRRM2 bound to TER S4 (PDB ID 4ERD),Citation13 the loop 4 is taken from the solution structure (PDB ID 2M21)Citation17 and placed on the distal stem 4.
Based on the p65 xRRM2-TER S4 complex, hLa RRM2 structures and the sequence analysis of 33 La and LARP7 proteins, we predicted that the xRRM motif might be found in the RRM2 of other La and LARP7 family proteins. Examination of the sequences of La and LARP7 showed that all La and LARP7 have a predicted secondary structure of an atypical RRM lacking RNP1 and RNP2 sequences, contain a helix α3 and residues C-terminal to α3 that could potentially form a helix α3x upon binding to RNA.Citation13,Citation25 From this analysis we also identified a new RNP3 sequence (F/Y/W)-X-(D/Q/E/N) on β2 and a conserved Arg (R465) on β3, which could recognize single-strand RNA nucleotides (see below), as they do for the GA bulge in TER S4 ().Citation13 Interestingly, the conserved Arg lies at the position on β3-strand where the canonical RNP1 motif is located (). Elucidation of the exact mode of RNA binding for these predicted xRRMs awaits future structural studies.Citation13
Unique features of the xRRM2 but not the long β2-β3 loop are important for RNA recognition
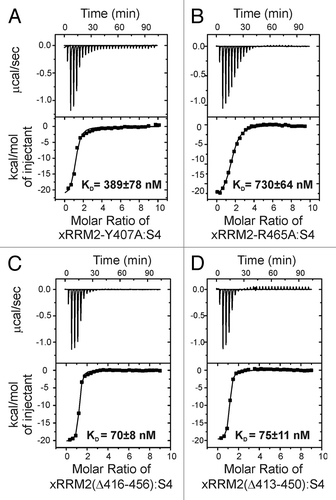
The structure of the complex of p65 xRRM2 with RNA revealed specific interactions between residues of the proposed RNP3 on β2 and the conserved R465 on β3 for recognition of single-stranded RNA (the GA bulge) and the C-terminal α3x for binding to the major groove of double-stranded RNA. We previously tested the importance of these interactions by individual alanine substitution of interacting residues and deletion of residues of α3x, by measuring KD using the crystal structure constructs of xRRM2 (xRRM2Δ413-459), in which the unusually long β2-β3 loop was completely deleted.Citation13 Here, we compare those results to the same mutations in the context of the WT xRRM2 (no β2-β3 loop deletion). The conserved RNP3 residue Y407 stacks on the G121 of the conserved GA bulge and its hydroxyl group has potential hydrogen bonds to the backbone at G121. Mutation of Y407A results in TER S4 interaction with KD of 389 ± 78 nM () and 375 ± 27 nMCitation13 for xRRM2-Y407A and xRRM2(Δ413-459)-Y407A, respectively, which is an ~11-fold decrease for both compared with WT xRRM2. The conserved R465 of p65 xRRM2 recognizes both G121 and A122 of TER S4 by forming specific hydrogen bonds to the Hoogsteen edge of these bases. Mutation of R465A results in TER S4 interaction with KD of 730 ± 64 nM () and 639 ± 47 nMCitation13 for xRRM2-R465A and xRRM2(Δ413-459)-Y407A, respectively, which is ~20-fold decrease for both compared with WT xRRM2 (). These results show that the contacts in the crystal structure construct are equally relevant in the context of the full-length WT xRRM2.
Figure 3. Isothermal titration calorimetry (ITC) data and analysis of xRRM2 mutants with TER S4. (A) TER S4 and p65 xRRM2-Y407A mutant (from RNP3). (B) S4 and p65 xRRM2-R465A (conserved R465 in β3). (C) TER S4 and p65 xRRM2(Δ416-456). (D) TER S4 and p65 xRRM2(Δ413-450). The KDs of interactions are shown on each panel.
Deletion of α3x from xRRM2 abolished the binding to TER S4, and deletion of α3x in the full-length p65 decreased binding to full-length TER and abolished the hierarchical assembly of TERT with p65-TER.Citation13 Interestingly, deletion of the conserved GA bulge in S4 decreases but does not completely abolish binding (KD = 2985 ± 220 nM vs. 36 ± 4 nM, ~80-fold decrease).Citation13 This indicates that p65 xRRM2 interacts with duplex RNA, probably via insertion of α3x into the major groove of RNA.
p65 xRRM2 has an unusually long β2-β3 loop that we deleted for the structural analysis. Although both domain folding and RNA binding were observed for p65 xRRM2 when the loop was deleted, we note that β2-β3 loops have been shown to be important for RNA or protein interactions for other RRMs.Citation23,Citation24,Citation27,Citation28 We tested the importance of the β2-β3 loop, which was partially (Δ422-442) and completely (Δ413-459) deleted in the NMR and crystal structures, respectively, for RNA binding. We have previously reported that p65 xRRM2 binds TER S4 RNA with a KD of 36.4 ± 3.6 nM.Citation13 Partial β2-β3 loop deletion in xRRM2(Δ422-442) does not decrease binding significantly (< 2 fold with KD = 50.3 ± 7.1 nM); however, complete deletion of β2-β3 loop residues in xRRM2(Δ413-459) decreases RNA binding by about 3-fold (KD = 101 ± 21.2 nM). Comparison of the position of the β2 and β3 strands in the solution structure of xRRM2(Δ422-442) (partial loop deletion) and crystal structure of xRRM2(Δ413-459) (complete loop deletion) showed small positional differences in the β2 and β3 strands, likely due to steric restraints from complete deletion of the loop. The positions of the β2 and β3 strands in the solution structure of the free crystal structure of xRRM2(Δ422-442) are closer to their positions in the xRRM2-S4 complex than those for free xRRM2(Δ413-459). We therefore attribute the 3-fold decrease in binding to RNA by xRRM2(Δ413-459) vs. xRRM2 to the consequent requirement for the β2 and β3 strands to move positions to interact with the RNA in the complex. To further test the importance of β2-β3 loop residues or length on RNA binding, we made two more loop truncation mutants xRRM2(Δ416-456) and xRRM2(Δ413-450), with 6 and 9 β2-β3 loop residues remaining, respectively, and measured their binding to RNA by ITC. The KDs of interaction of xRRM2(Δ416-456) and xRRM2(Δ413-450) for TER S4 was 70 ± 8 nM and 75 ± 11 nM, respectively (). These values are < 2-fold weaker compared with native xRRM2, similar to the result with xRRM2(Δ422-442) (15 residue β2-β3 loop).Citation13 Hence, we conclude that the β2-β3 loop does not play a significant role in RNA binding or domain folding. The β2-β3 loop has an overall slightly negative charge, an observation that is consistent with a potential role in protein rather than RNA binding. We previously showed that deletion of the β2-β3 loop has no effect on hierarchical assembly of the Tetrahymena telomerase catalytic core, indicating that there is no essential interaction with TERT. Sequence alignment of p65 with other La and LARP7 proteins did not predict an unusually long β2-β3 loop in other proteins.Citation13 Thus, a long β2-β3 loop is not a conserved feature of the xRRM.
Possible mechanism of RNA binding by xRRM2 of La and LARP7 proteins
The region C-terminal to the La module in genuine La and LARP7 proteins has been implicated in RNA chaperone and/or folding functions.Citation13,Citation29-Citation31 This part of the protein contains what we have identified as the xRRM2 ().Citation13,Citation31 How the xRRM2 interacts with RNA and whether its RNA folding functions are associated with the La module in genuine La and LARP7 proteins in general is not clear. The C-terminal extension includes important functional elements in hLa protein, such as the nuclear localization signal (NLS), a short basic motif (SBM), a nuclear retention element (NRE) and several phosphorylation sites.Citation2,Citation25 Recently, in an in vitro FRET based assay, the C-terminal region of hLa, which contains the predicted xRRM2 and additional residues was shown to have RNA strand annealing and strand dissociation activity associated with RNA chaperones.Citation29
A well-known and characterized target of hLa is hepatitis C virus (HCV) IRES mRNA.Citation32,Citation33 Recently, it was shown that the C-terminal half of hLa in concert with La module is required for HCV domain IV binding.Citation30 Both single-strand as well as structured duplex sequences were required for the association of hLa with RNA and the xRRM2 was shown to be critical for RNA binding, but the exact RNA sequence and structure requirements are still not clear. It appears that in genuine La proteins, the xRRM2 is important for RNA chaperoning activity; however, this function of xRRM2 in genuine La appears to be dependent on the La module.
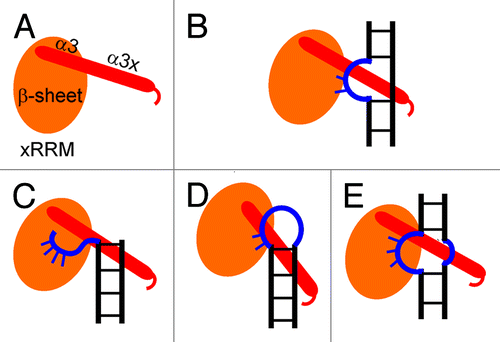
Based on all these observations and the structure of p65 xRRM2:TER S4 complex, we predict that other La and LARP7 xRRM2 will interact with RNA targets containing single-stranded RNA linked to double-stranded RNA. In all cases, the single-stranded RNA would be predicted to interact with the “front face” of the β-sheet using residues from RNP3 and the conserved Arg and other possible solvent-exposed residues and the α3x will bind a widened major groove near the single-stranded region (). Like the RNP1 and RNP2 on canonical RRMs, we predict that the RNP3 [(F/Y/W)-X-(D/Q/E/N)] on xRRM will be important for binding to one or more of the substrate nucleotides. The F/Y/W residue potentially provides stacking interactions and the D/Q/E/N residue provides hydrogen bonds to RNA nucleotides, analogous to Y407 and D409 in p65 xRRM2. This combined single- and double-strand RNA-binding mode of interaction by xRRM2 of genuine La and LARP7 proteins can be employed for diverse RNA substrates. Asymmetric or symmetric internal loops, apical loops or single-strand overhangs could provide the single-strand RNA for interaction with the β-sheet surface and the α3x could bind the double-strand RNA near the single stranded regions (). Thus, just as canonical RRMs can recognize diverse lengths and sequences of single-stranded RNA, the xRRM can be specific for a variety of secondary structures combining single- and double-strand RNA. Future studies of predicted xRRMs from other LARP7 and genuine La proteins are required to test this hypothesis.
Figure 4. Proposed binding modes for recognition of different RNAs by the xRRM. (A) Cartoon depiction of xRRM, the β-sheet is shown in orange oval and helix 3 (α3+α3x) is shown in red cylinder based on p65 xRRM2. (B) Cartoon depiction of p65 xRRM2 and TER S4 interaction. (C‒E) Different scenarios of single- and double-strand RNA interaction mode for xRRM2 of La and LARP7 protein.
Does RRM1 in La and LARP7 have RNA chaperone activities similar to xRRM2?
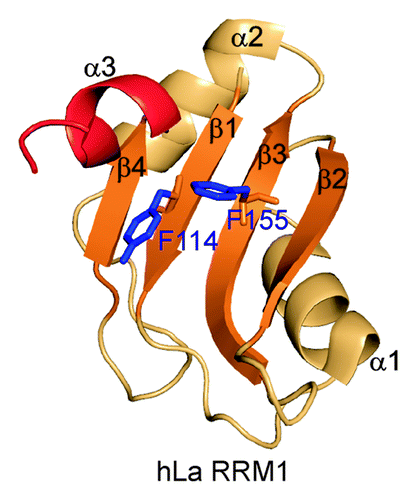
Although the La module is involved in UUU-3′OH recognition, this interaction leaves the canonical RNA binding “front face” of RRM1 available for RNA interaction ().Citation34-Citation36 The hLa RRM1 is an atypical RRM with only partial conservation of RNP1 and RNP2 motifs and an α3 at its C terminus. However, the α3 does not lie across the surface of the upper half of the β-sheet, but rather protrudes out almost perpendicular to the β-sheet ().Citation13,Citation34-Citation36 It has been proposed that the RRM1 of La and LARP7 proteins may have RNA chaperone functions, in addition to UUU-3′OH recognition, which may be analogous to the p65 xRRM2 RNA binding and folding functions. Some recent studies support this hypothesis. In the recent FRET-based study, the authors reported that the N-terminal half of hLa that includes the La module also had a robust RNA chaperone activity independent of its UUU-3′OH binding function and of xRRM2.Citation29 The non-canonical helix α3 after the RRM1 was shown to be critical for the RNA chaperoning functions. Fusion of RRM1 α3 to U1A protein imparted an RNA chaperoning function to the chimeric U1A-α3 protein.Citation29 In the same paper, the authors showed that mutation of the conserved RNP2 (β1) residue Y114A and RNP1 (β3) residue F155A resulted in loss of RNA chaperone functions.Citation29 These key residues in both human and yeast La protein had previously been shown to be important for tRNA binding and rescue of tRNA-mediated suppression.Citation18
Figure 5. Structure of the hLa RRM1 (PDB ID 2VOP). All the secondary structure elements are marked. RNP1 and RNP2 residues 114 and 155, respectively, and non-canonical helix α3 are marked.
Yeast genuine La (Lhp1p) does not have a C-terminal RRM2 domain; however, it has a C-terminal unstructured extension after RRM1 that is required for several Lhp1p functions.Citation1,Citation31 This region of the protein gets structured and protease resistant upon binding to certain structured non-coding RNA precursors, including pre-tRNA.Citation31 Based on these observations, it appears that the single RRM of Lhp1p may be an xRRM and play a dual role in protecting RNA from degradation by binding the UUU-3′OH and as a chaperone for folding structured RNAs in a manner similar to p65 xRRM2. Further, it is possible that hLa has RNA chaperone functions associated with both an xRRM-like RRM1 and xRRM2.
Role of telomerase La family protein p65 in telomerase biogenesis and assembly
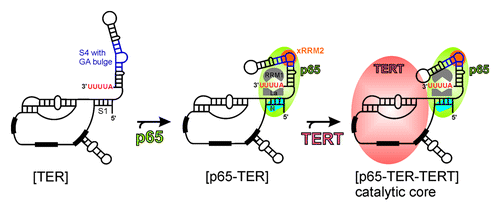
LARP7s are distinct from genuine La proteins in that besides UUU-3′OH binding, they have distinct and specific RNA binding and chaperone activities and they are stable components of the target RNPs.Citation2,Citation3 p65 is thought to bind the UUU-3′OH of TER as soon as it is transcribed by RNAPIII, protecting it from exonucleolytic degradation (). Since Tetrahymena TER is not further processed to remove the UUU-3′OH, p65 remains associated with TER, where it plays an essential role in vivo in RNP assembly. A model for hierarchical assembly of p65-TER-TERT catalytic core is shown in . Based on the existing literature and our work we propose that p65 will bind the TER 3′ AUUUU tail in a manner similar to hLa and 3′ UUU-3′OH interactions. This will position xRRM2 to bind the GA bulge in the SL4. Binding of xRRM2 to SL4 causes conformational changes in TER, which organize the RNA for TERT binding.Citation13
Figure 6. Model of assembly of Tetrahymena telomerase catalytic core. The known and proposed binding sites for different domains of p65 to TER are shown. Facilitation of TERT binding to p65-TER complex leading to hierarchical catalytic core assembly is shown. The conserved secondary structural elements of TER are marked in .
Materials and Methods
RNA and protein preparation
xRRM2 loop deletion mutants xRRM2(Δ422-442) and xRRM2(Δ413-459) and RNP3 mutants xRRM2-Y407A and xRRM2-R465A were made using side-directed mutagenesis as per the vendor instructions (Agilent Technologies). Mutations were verified by DNA sequencing. Both protein and S4 RNA were prepared as previously described.Citation13 S4 is the minimal stem 4 TER RNA construct that contains four base pairs on either side of the GA bulge and is capped by a UUCG tetraloop. S4 was used in our previous structural studies.Citation13 The integrity and purity of proteins and RNA were checked on denaturing polyacrylamide gels (PAGE) and by NMR (data not shown).
Isothermal calorimetry (ITC) titration
Interaction between xRRM2 mutants and TER S4 RNA was measured by ITC using a MicroCal Omega VP-ITC system (MicroCal). Both RNA and protein were dialyzed against 20 mM NaH2PO4, 50 mM NaCl, pH 7 buffer prior to titration. Five micromolar RNA in the cell was titrated with 225 μM protein using VP-ITC machine (MicroCal). Data was analyzed using ORIGIN v5.0 (MicroCal). The binding parameters ΔH, association constant K and n were kept floating during the fits. The average heat change from the last four injections was subtracted from the data to account for the heat of dilution.
| Abbreviations: | ||
| TER | = | telomerase RNA |
| TERT | = | telomerase reverse transcriptase |
| LARP7 | = | La-related protein 7 |
| RRM | = | RNA recognition motif |
| RNAPIII | = | RNA polymerase III |
| ITC | = | isothermal titration calorimetry |
| WT | = | wild-type |
Acknowledgments
This work was supported by NSF grant MCB1022379 and NIH grant GM48123 to J.F.
References
- Wolin SL, Cedervall T. The La protein. Annu Rev Biochem 2002; 71:375 - 403; http://dx.doi.org/10.1146/annurev.biochem.71.090501.150003; PMID: 12045101
- Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta 2010; 1799:365 - 78; http://dx.doi.org/10.1016/j.bbagrm.2010.01.011; PMID: 20138158
- Bousquet-Antonelli C, Deragon JM. A comprehensive analysis of the La-motif protein superfamily. RNA 2009; 15:750 - 64; http://dx.doi.org/10.1261/rna.1478709; PMID: 19299548
- Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 2011; 3; http://dx.doi.org/10.1101/cshperspect.a003558; PMID: 20660025
- Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA 2012; 18:1747 - 59; http://dx.doi.org/10.1261/rna.034629.112; PMID: 22875809
- Podlevsky JD, Chen JJ. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutat Res 2012; 730:3 - 11; http://dx.doi.org/10.1016/j.mrfmmm.2011.11.002; PMID: 22093366
- Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol 2006; 7:484 - 94; http://dx.doi.org/10.1038/nrm1961; PMID: 16829980
- Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev 2004; 18:1107 - 18; http://dx.doi.org/10.1101/gad.1201704; PMID: 15131081
- Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell 2009; 36:609 - 19; http://dx.doi.org/10.1016/j.molcel.2009.09.041; PMID: 19941821
- O’Connor CM, Collins K. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol 2006; 26:2029 - 36; http://dx.doi.org/10.1128/MCB.26.6.2029-2036.2006; PMID: 16507983
- Prathapam R, Witkin KL, O’Connor CM, Collins K. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol 2005; 12:252 - 7; http://dx.doi.org/10.1038/nsmb900; PMID: 15696174
- Stone MD, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 2007; 446:458 - 61; http://dx.doi.org/10.1038/nature05600; PMID: 17322903
- Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell 2012; 47:16 - 26; PMID: 22705372
- Akiyama BM, Loper J, Najarro K, Stone MD. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA 2012; 18:653 - 60; http://dx.doi.org/10.1261/rna.031377.111; PMID: 22315458
- Berman AJ, Gooding AR, Cech TR. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol 2010; 30:4965 - 76; http://dx.doi.org/10.1128/MCB.00827-10; PMID: 20713447
- Robart AR, O’Connor CM, Collins K. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA 2010; 16:563 - 71; http://dx.doi.org/10.1261/rna.1936410; PMID: 20106956
- Richards RJ, Wu H, Trantirek L, O’Connor CM, Collins K, Feigon J. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA 2006; 12:1475 - 85; http://dx.doi.org/10.1261/rna.112306; PMID: 16809815
- Bayfield MA, Maraia RJ. Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct Mol Biol 2009; 16:430 - 7; http://dx.doi.org/10.1038/nsmb.1573; PMID: 19287396
- Aigner S, Postberg J, Lipps HJ, Cech TR. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 2003; 42:5736 - 47; http://dx.doi.org/10.1021/bi034121y; PMID: 12741831
- Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol 2009; 6:122 - 8; http://dx.doi.org/10.4161/rna.6.2.8115; PMID: 19246988
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 2008; 29:588 - 99; http://dx.doi.org/10.1016/j.molcel.2008.01.003; PMID: 18249148
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem 2012; 81:119 - 43; http://dx.doi.org/10.1146/annurev-biochem-052610-095910; PMID: 22404626
- Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 2008; 18:290 - 8; http://dx.doi.org/10.1016/j.sbi.2008.04.002; PMID: 18515081
- Muto Y, Yokoyama S. Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems. Wiley Interdiscip Rev RNA 2012; 3:229 - 46; http://dx.doi.org/10.1002/wrna.1107; PMID: 22278943
- Jacks A, Babon J, Kelly G, Manolaridis I, Cary PD, Curry S, et al. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure 2003; 11:833 - 43; http://dx.doi.org/10.1016/S0969-2126(03)00121-7; PMID: 12842046
- Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J 2006; 25:3156 - 66; http://dx.doi.org/10.1038/sj.emboj.7601195; PMID: 16778765
- Hom RA, Chang PY, Roy S, Musselman CA, Glass KC, Selezneva AI, et al. Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain. J Mol Biol 2010; 400:145 - 54; http://dx.doi.org/10.1016/j.jmb.2010.04.067; PMID: 20460131
- Skrisovska L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, et al. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep 2007; 8:372 - 9; http://dx.doi.org/10.1038/sj.embor.7400910; PMID: 17318228
- Naeeni AR, Conte MR, Bayfield MA. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. J Biol Chem 2012; 287:5472 - 82; http://dx.doi.org/10.1074/jbc.M111.276071; PMID: 22203678
- Martino L, Pennell S, Kelly G, Bui TT, Kotik-Kogan O, Smerdon SJ, et al. Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res 2012; 40:1381 - 94; http://dx.doi.org/10.1093/nar/gkr890; PMID: 22009680
- Kucera NJ, Hodsdon ME, Wolin SL. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc Natl Acad Sci USA 2011; 108:1308 - 13; http://dx.doi.org/10.1073/pnas.1017085108; PMID: 21212361
- Pudi R, Abhiman S, Srinivasan N, Das S. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J Biol Chem 2003; 278:12231 - 40; http://dx.doi.org/10.1074/jbc.M210287200; PMID: 12540850
- Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol 2004; 24:6861 - 70; http://dx.doi.org/10.1128/MCB.24.15.6861-6870.2004; PMID: 15254251
- Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR, Curry S. Structural analysis reveals conformational plasticity in the recognition of RNA 3′ ends by the human La protein. Structure 2008; 16:852 - 62; http://dx.doi.org/10.1016/j.str.2008.02.021; PMID: 18547518
- Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, et al. Structural basis for recognition and sequestration of UUU(OH) 3′ termini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell 2006; 21:75 - 85; http://dx.doi.org/10.1016/j.molcel.2005.10.027; PMID: 16387655
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, et al. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 2004; 11:323 - 9; http://dx.doi.org/10.1038/nsmb747; PMID: 15004549