Abstract
At low temperatures the Escherichia coli rpoS mRNA, encoding the stationary phase sigma factor RpoS, forms an intramolecular secondary structure (iss) that impedes translation initiation. Under these conditions the small RNA DsrA, which is stabilzed by Hfq, forms a duplex with rpoS mRNA sequences opposite of the ribosome-binding site (rbs). Both the DEAD box helicase CsdA and Hfq have been implicated in DsrA·rpoS duplex formation. Hfq binding to A-rich sequences in the rpoS leader has been suggested to restructure the mRNA, and thereby to accelerate DsrA·rpoS duplex formation, which, in turn, was deemed to free the rpoS rbs and to permit ribosome loading on the mRNA. Several experiments designed to elucidate the role of Hfq in DsrA-mediated translational activation of rpoS mRNA have been conducted in vitro. Here, we assessed RpoS synthesis in vivo to further study the role of Hfq in rpoS regulation. We show that RpoS synthesis was reduced when DsrA was ectopically overexpressed at 24 °C in the absence of Hfq despite of DsrA·rpoS duplex formation. This observation indicated that DsrA·rpoS annealing may not be sufficient for efficient ribosome loading on rpoS mRNA. In addition, a HfqG29A mutant protein was employed, which is deficient in binding to A-rich sequences present in the rpoS leader but proficient in DsrA binding. We show that DsrA·rpoS duplex formation occurs in the presence of the HfqG29A mutant protein at low temperature, whereas synthesis of RpoS was greatly diminished. RNase T1 footprinting studies of DsrA·rpoS duplexes in the absence and presence of Hfq or HfqG29A indicated that Hfq is required to resolve a stem-loop structure in the immediate coding region of rpoS mRNA. These in vivo studies corroborate the importance of the A-rich sequences in the rpoS leader and strongly suggest that Hfq, besides stabilizing DsrA and accelerating DsrA·rpoS duplex formation, is also required to convert the rpoS mRNA into a translationally competent form.
Keywords: :
Introduction
In bacteria, small regulatory trans-acting RNAs (sRNAs) can modulate different stress responses through post-transcriptional regulation.Citation1,Citation2 These sRNAs either prevent ribosome loading onto the mRNA by base-pairing with or in the vicinity of the ribosome binding site (rbs) or activate translation by abrogating intramolecular inhibitory stem-loop structures (iss) that block ribosome binding. The RNA chaperone Hfq has been shown to be crucial for riboregulation in Enterobacteriaceae, which results on the one hand from binding to and protection of sRNAs from nucleolytic decay,Citation3-Citation5 and on the other hand from accelerating annealing of sRNAs with their target mRNAs.Citation3 E. coli Hfq hexamers have dedicated RNA binding sites, preferably binding uridine-rich stretches of sRNAs around the central pore of the proximal surfaceCitation6-Citation10 and A-rich sequences on the distal surface.Citation11 Moreover, binding of the sRNA RybB to Hfq not only involves contacts via 3′ terminal U-rich stretches but also with the lateral surface of the hexamer.Citation12 The dedicated sRNA and mRNA binding surfaces on either site of the Hfq-hexamer may serve to transiently increase the local concentration of two RNA substrates. In addition, the inherent capacity of Hfq to induce conformational changes in RNAs,Citation13-Citation15 together with the observed structural flexibility of RNA ligands bound to Hfq,Citation8,Citation10 may stochastically facilitate base-pairing.
A paradigm for Hfq-mediated translational activation by a sRNA is the E. coli rpoS mRNA, encoding the stationary phase sigma factor RpoS. The sRNA DsrA, which is mainly transcribed at low-growth temperatureCitation16,Citation17 (≤ 25 °C), is necessary for translational activation of rpoS mRNACitation18 under these conditions. The DsrA-rpoS mRNA interaction counteracts an iss that impedes ribosomal access to the rbs of rpoS.Citation19 In vivo DsrA·rpoS duplex formation at low temperature further requires the CsdA helicase,Citation20 and creates RNase III cleavage sites within the duplex that prevents reuse of DsrA.Citation21 DsrA was shown to bind with a 1:1 stoichiometry on the proximal face of Hfq,Citation8,Citation22 whereas rpoS mRNA is recruited to the distal side of Hfq via A-rich elements in the mRNA leader.Citation23,Citation24 The interaction of Hfq with both RNAs, albeit accelerating annealing, appears to be transient.Citation23,Citation25 The need for Hfq to cycle off its binding site on DsrA prior to or during annealing with rpoSCitation26 could result from the fact that at least part of the Hfq binding site on DsrA base-pairs with rpoS mRNA.Citation27,Citation28 Moreover, Hfq binding to an (AAN)4 repeat element in the rpoS leader has been shown to be critical for regulation by sRNAs,Citation24 and for DsrA·rpoS duplex formation.Citation29 Binding to these A-rich motifs has been suggested to result in restructuring of the rpoS leader, priming the mRNA for productive interactions with DsrA.Citation29
Despite intensive research on the DsrA-rpoS model system, the Hfq-dependent steps toward ribosome loading on rpoS mRNA at low temperature are not completely understood. Here, we have addressed in vivo whether DsrA·rpoS annealing is sufficient for efficient RpoS synthesis at low temperature. Although DsrA·rpoS duplex occurred in the absence of Hfq as well as in the presence of a HfqG29A mutant protein deficient in binding to the (AAN)4 motifs, RpoS synthesis was diminished. RNase T1 footprinting studies of DsrA·rpoS duplexes in the absence and presence of Hfq or HfqG29A indicated that Hfq is required to resolve a stem-loop structure in the immediate coding region of rpoS mRNA. These in vivo studies corroborate the importance of the A-rich sequences in the rpoS leader and show that Hfq, besides accelerating DsrA·rpoS duplex formation, is apparently additionally required to convert the rpoS mRNA into a translationally competent form.
Results and Discussion
Hfq- and DsrA-independent synthesis of RpoS at 37 °C
The design of in vivo studies to assess the requirement for Hfq in rpoS translation is complicated as Hfq performs several interdependent functions; (1) it affects the stability of sRNAs involved in rpoS translational regulation, e.g., DsrA,Citation4,Citation18,Citation24 (2) it accelerates the interaction between DsrA and rpoS mRNA,Citation25 and (3) it has been implicated in restructuring the rpoS leader, which, in turn, is deemed to be required for DsrA·rpoS duplex formation.Citation29 In addition, some studies addressing the requirement for sRNAs and/or Hfq were performed at 37 °C,Citation24 where DsrA is less abundant than at 24 °CCitation16,Citation17 (, lanes 2 and 4). In this study, we aimed at dissecting Hfq and/or DsrA-dependent steps that lead to in vivo RpoS synthesis. First, we tested whether the temperature affects RpoS synthesis in a Hfq-dependent manner. The hfq- strain JW4130 was transformed with plasmids pACYC184 (control) or pAHfq (encodes Hfqwt) and cultivated at 24 °C and 37 °C, respectively. As expected, no RpoS synthesis was observed in the absence of DsrA and Hfq at 24 °C in strain JW4130(pACYC184), whereas RpoS synthesis occurred in strain JW4130(pAHfq) in the presence of DsrA and Hfq, respectively (, lanes 1 and 2). Most likely, the absence of DsrA in strain JW4130(pACYC184) is attributable to a lack of stabilization by Hfq.Citation4,Citation18,Citation24 In contrast, RpoS synthesis was rather independent of Hfq and DsrA at 37 °C. The RpoS levels in strain JW4130(pACYC184) equalled ~66% of that observed in strain JW4130(pAHfq) (, lanes 3 and 4). Although the presence of Hfq and DsrA stimulated translation of rpoS mRNA in strain JW4130(pAHfq) at 37 °C, both are apparently not essential, i.e., the iss occluding the ribosome binding site of rpoS appears to be not stable enough to efficiently counteract initiating ribosomes at this temperature. It seems worth noting that this can lead to false-positive results when Hfq mutant proteins are tested for their capacity to stimulate RpoS translation at 37 °C.
Figure 1. Hfq/DsrA requirement at 24 °C and 37 °C. The steady-state levels of RpoS were determined by quantitative western blotting in the E. coli hfq- strain JW4130 harboring the control plasmid pACYC184 (lanes 1 and 3) and plasmid pAHfq, encoding Hfq (lanes 2 and 4), respectively. The strains were grown to early log phase (OD600 of 0.4) at 24 °C (lanes 1 and 2) or 37 °C (lanes 3 and 4). Equal amounts of cellular protein were loaded onto the SDS-polyacrylamide gel. Immunodetection of RpoS, ribosomal protein L14 (loading control), and Hfq as well as the detection of DsrA and 5S rRNA (loading control) by northern blot analysis was performed as described in the Materials and Methods. Only the relevant parts of the immunoblots and the autoradiographs are shown.
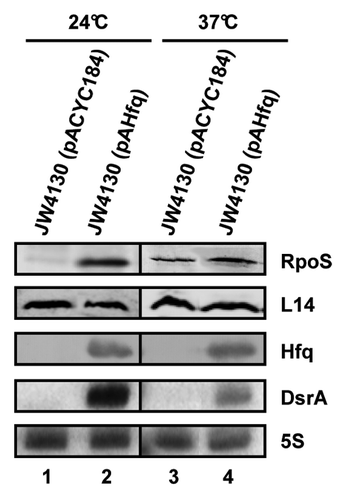
Similarly, Soper et al.Citation24 showed that a rpoS::lacZ translational fusion gene is translated at 37 °C with an approximately 8-fold higher rate in a hfq+ strain when compared with a hfq-- strain. In addition to DsrA, two other sRNAs, RprACitation30 and ArcZ,Citation31 are involved in translational activation of rpoS mRNA. As these sRNAs are likewise destabilized in a hfq-- background,Citation24 and as the activity of a rpoS::lacZ fusion was higher in a triple dsrA, rprA, and arcZ mutant background than in a hfq deletion mutant at 37 °C,Citation31 the observed difference in RpoS synthesis between the hfq- strain and the hfq+ strain at 37 °C (, lanes 3 and 4) could be attributable to the function of Hfq rather than to that of the sRNAs. Clearly, we cannot disregard the possibility that the observed differences in RpoS/RpoS-LacZ synthesis result from a reduced stability of the mRNA at 37 °C in the absence of Hfq. However, this seems less likely, as no differences in the steady-state levels of full-length rpoS mRNA were previously observed in the presence and absence of Hfq at 37 °C.Citation21
DsrA·rpoS duplex formation occurs in the absence of Hfq at 24 °C
To test whether DsrA·rpoS duplex formation is the decisive event or whether the RNA chaperone function of Hfq stimulates, besides its effects on DsrA stability and DsrA·rpoS pairing, efficient translation of rpoS at low temperature, we next asked whether over-production of DsrA at 24 °C can compensate for the absence of Hfq. Overexpression of dsrA resulted in increased levels of DsrA in the hfq- when compared with the hfq+ background, wherein hfq was ectopically expressed from plasmid pAHfq and DsrA originated from the endogenous gene, respectively (, lanes 2 and 4). When compared with the hfq- and DsrA-overexpressing strain JW4130(pNM13), RpoS synthesis was ~7-fold increased in the hfq+ strain JW4130(pAHfq) (, lanes 2 and 4).
Figure 2. DsrA·rpoS duplex formation does not lead to efficient translation of rpoS mRNA in the absence of Hfq. (A) Immunodetection of RpoS, L9 ribosomal protein (loading control), and Hfq protein and detection of DsrA and 5S rRNA in the hfq- strain JW4130 harboring plasmid pNM12 (control; lane 1), pNM13 (encoding DsrA; lane 2), pACYC184 (control; lane 3), and pAHfq (encoding hfq; lane 4), respectively. The proteins and RNAs were visualized as described in the legend to . Only the relevant sections of the immunoblots and autoradiographs are shown. (B) Lanes 1–4, primer extension analysis of total RNA isolated from strains JW4130(pNM12), JW4130(pNM13), JW4130(pACYC184), and JW4130(pAHfq), respectively. The primer extension (PE) signals for rpoS mRNA isolated from the different strains are shown on top. The RNase III-mediated cleavage signals in rpoS mRNA are marked by arrows (G-112 and A-109). U, A, C, G, sequencing ladder.
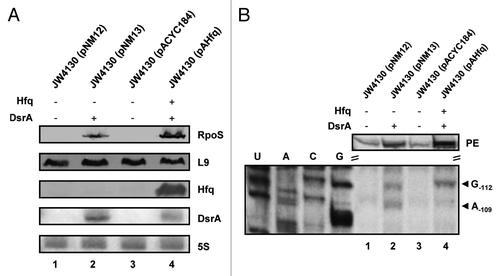
To test whether the reduced synthesis of RpoS could result from diminished DsrA·rpoS duplex formation in the hfq- strain JW4130(pNM13), we utilized the reported RNase III specific cleavages of rpoS mRNA upon duplex formation with DsrACitation21,Citation32 (Fig. S1) as a diagnostic marker. The strains JW4130(pMN12), JW4130(pMN13), JW4130(pACYC184), and JW4130(pAHfq) were grown to an OD600 of 0.4 at 24 °C, when total RNA was purified from either strain. RNase III cleavage of rpoS mRNA was assessed by primer extension using a [32P]-5′end-labeled rpoS-specific primer. As anticipated, no RNase III specific cleavage signals were detected at positions -112Citation21 and -109Citation32 of rpoS mRNA in the absence of DsrA and Hfq (, lanes 1 and 3). Notably, the levels of full-length rpoS mRNA were reduced in the absence of Hfq and DsrA in strains JW4130(pMN12) and JW4130(pACYC184), respectively (, lanes 1 and 3; PE). This can be reconciled with a faster turnover of un-translated rpoS mRNA in the absence of DsrA, which has been attributed to initial RNase III clevages at positions -94 and -15 in the 5′UTR of rpoS mRNA.Citation21 In support, mutations in dsrA that disrupt pairing between DsrA and rpoS also destabilize the mRNA.Citation32. In contrast, alternative cleavage sites at positions -112Citation21 and -109Citation32 of rpoS mRNA are created upon DsrA·rpoS pairing, which contributes to translation and furthermore to stabilization of rpoS mRNA.Citation21
RNase III dependent cleavage signals at positions -112 and -109 in rpoS mRNA were observed in both, the hfq- strain as well as in the hfq+ strain in the presence of DsrA (, lanes 2 and 4), showing that Hfq is not essential for DsrA·rpoS duplex formation at low temperatures. When compared with strain JW4130(pNM13), RpoS synthesis was increased in the hfq+ background JW4130(pAHfq) (, lanes 2 and 4). However, as apparent from the full-length primer extension signals (, lanes 2 and 4), the steady-state levels of rpoS mRNA were as well somewhat increased in JW4130(pAHfq) when compared with the hfq- and DsrA-overexpressing strain JW4130(pNM13). We interpreted this as showing that the increased synthesis of RpoS in strain JW4130(pAHHfq) leads to stabilization of rpoS mRNA.Citation21,Citation32 Nevertheless, it should be noted that the mRNA levels in JW4130(pAHfq) were only ~1.5-fold higher than in JW4130(pNM13), whereas the RpoS levels were ~7-fold higher in JW4130(pAHfq) than in JW4130(pNM13). In addition, the intensity of the RNase III-dependent cleavage signals at positions -112 and -109 of rpoS mRNA indicated more rpoS mRNA in complex with DsrA in strain JW4130(pNM13) than in strain JW4130(pAHfq). Hence, these studies suggested that DsrA·rpoS duplex formation may not suffice for efficient translation of rpoS mRNA, and that Hfq, besides stabilizing DsrACitation4,Citation18,Citation24 and facilitating pairing between both RNAs,Citation25 positively affects the translatability of rpoS mRNA at low temperature.
DsrA·rpoS duplex formation is not sufficient for translation initiation of rpoS mRNA
To further study whether Hfq is per se stimulating efficient translation at low temperatures, we made use of a Hfq mutant deficient in binding to the A-rich motifs present in the rpoS leader, but proficient in DsrA binding. The distal face of the Hfq hexamer harbors six tripartite binding motifs that consist of an adenosine-specific site (A-site), a purine nucleotide selective site (R-site), and a sequence-non-discriminating RNA entrance/exit site (E-site).Citation11 Common to all binding sites is the coordination of the adenosine ribose 2’-OH by the carbonyl-group of Gly-29.Citation33 This coordination was reported to likely convey selectivity for RNA over DNA for this binding site.Citation11 Gly-29 adopts main-chain conformation allowing only for residues, which do not bare a side-chain. This suggested that any substitution of Gly-29 will dramatically reduce the capacity of Hfq to bind an adenosine ligand in the R-site. We therefore constructed a hfqG29A mutant allele with the aim to reduce binding of HfqG29A to the A-rich motifs present in the rpoS leader but capable to bind to DsrA. As anticipated, purified HfqG29A did not bind to the (AAN)4 repeat (Fig. S2), but retained the ability to bind to DsrA (Fig. S3). Accordingly, HfqG29A stabilized DsrA in vivo as the levels of the sRNA were comparable in the presence of Hfqwt and HfqG29A (, lanes 1 and 2).
Figure 3. DsrA·rpoS duplex formation in the presence of HfqG29A does not result in efficient translation of rpoS mRNA at low temperature. (A) Immunodetection of RpoS, ribosomal protein L9, and Hfq in strains JW4130(pAHfq) (lane 1), JW4130(pAHfqG29A) (lane 2), and JW4130(pACYC184) (lane 3). DsrA and 5S rRNA were detected as described in the legend to . Only the relevant parts of the immunoblots and autoradiographs are shown. (B) Lanes 1–3, primer extension analysis of total RNA isolated from strains JW4130(pAHfq), JW4130(pAHfqG29A), and JW4130(pACYC184), respectively. The primer extension (PE) signals for rpoS mRNA isolated from the different strains are shown on top. The RNase III-mediated cleavage signals in rpoS mRNA are marked by arrows (G-112 and A-109). G, G sequencing ladder.
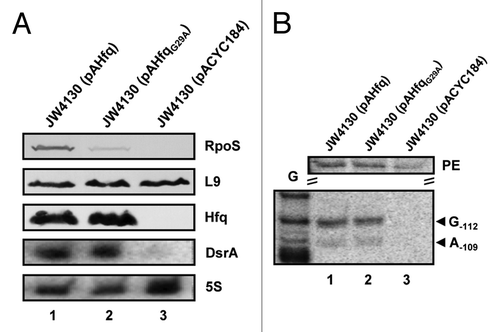
Next, we tested whether the apparent deficiency of HfqG29A in binding to the (AAN)4 motif affects DsrA·rpoS duplex formation at 24 °C by assessing RNase III processing of rpoS mRNA in strains JW4130(pAHfq), JW4130(pAHfqG29A), and JW4130(pACYC184), respectively. In the absence of Hfq/DsrA, the rpoS leader was not cleaved at positions -112/-109 in strain JW4130(pACYC184) (, lane 3). The intensities of the RNase III-dependent cleavage signals obtained in strains JW4130(pAHfq) and JW4130(pAHfqG29A) were comparable (, lanes 1 and 2), showing that DsrA·rpoS duplex formation occurs in the presence of HfqG29A under these conditions at low temperature. Nevertheless, despite almost equal steady-state levels of full-length rpoS mRNA (, lanes 1 and 2; PE) in JW4130(pAHfq) and JW4130(pAHfqG29A), RpoS was hardly synthesized in the presence of HfqG29A in strain JW4130(pAHfqG29A) (, lane 2). This observation suggested that binding of Hfq to the rpoS leader may, in addition to accelerating DsrA·rpoS duplex formation,Citation23,Citation25 restructure the rpoS mRNA into a translationally competent form.
To support this hypothesis, structural probing with RNase T1 was performed in vitro on DsrA·rpoS duplexes in the absence of Hfq and in the presence of Hfqwt or HfqG29A protein. In vitro synthesized full-length rpoS mRNA was first annealed to full-length DsrA and a [32P]-5′-end labeled primer. Upon cooling to 24 °C, the annealing mix was incubated for 10 min in the absence of Hfq and in the presence of Hfqwt or HfqG29A, followed by incubation with RNase T1. Next, reverse transcriptase was added to map the T1-dependent cleavage signals. Most obvious was the difference in the cleavage pattern observed in the presence and absence of Hfqwt within and adjacent to the rpoS start codon. In the absence of Hfqwt (, lane 2) cleavage at position G+3, G+5, and G+9 was hardly obvious. In addition, pronounced stop signals appeared at position A+1 and A+8. In contrast, in the presence of Hfqwt, G+3, G+5, and G+9 were accessible for RNase T1 and the stop signals were not observed (, lane 3). Therefore, the reduced accessibility of the start codon for productive interactions with initiator tRNA could explain why rpoS mRNA is not efficiently translated at 24 °C in the absence of Hfq but presence of DsrA (, lane 2). Moreover, as G+3, G+5, and G+9 were protected in the absence of Hfqwt, whereas the G+15, G+18, and G+22 were accessible for RNase T1, the cleavage pattern indicated the presence of a local secondary structure including the G+3 of the start codon and sequences immediately downstream (Fig. S4). Thus, in addition to the reduced accessibility of the start codon, the local secondary structure could as well interfere with ribosome binding in the absence of Hfq. At present, we can also not exclude Hfq-induced conformational changes further upstream, which might impact on rpoS translation. Although RNase III cleavage of the DsrA·rpoS duplex was omitted in these structural probing experiments, it seems less likely that cleavage at positions -112Citation21 and -109Citation32 in rpoS mRNA would affect this local secondary structure. Taken together, these results would support the notion that DsrA·rpoS duplex formation is not sufficient for efficient ribosome loading on rpoS mRNA, and that restructuring by Hfq is also required to convert rpoS mRNA into a translationally competent form. Even though the accessibility of G+3, G+5, and G+9 for RNase T1 increased as well in the presence of HfqG29A, deprotection was not as pronounced as with Hfqwt. In addition, in contrast to Hfqwt, the stop signal at A+8 was maintained in the presence of HfqG29A (, lane 4). Thus, the lack of HfqG29A binding to the (AAN)4 motif in the rpoS leader could lead to translationally incompetent populations of rpoS mRNA, which, in turn, could explain its deficiency in rpoS translation (, lane 2).
Figure 4. Structural probing of the translation initiation region of rpoS mRNA in complex with DsrA in the absence of Hfq and in the presence of Hfqwt or HfqG29A. First, rpoS mRNA was annealed to DsrA and to the [32P]-5′-end labeled primer. The annealing mix was cooled to 24 °C and divided into three parts followed by incubation for 10 min in the absence and presence of Hfqwt and HfqG29A, respectively. Then RNase T1 was added for 10 min followed by primer extension with AMV reverse transcriptase at 45 °C. The primer extension products were separated on a 8% polyacrylamide-8M urea gel and visualized by a PhosphorImager (Molecular Dynamics). The sequence of the immediate coding region is shown at the left. The start codon is marked by dots. RNase T1 cleavage 3′ of G residues and stop signals of the reverse transcriptase at A residues are indicated at the right. The numbering is given with regard to the A of the start codon (+1). Lane 1, primer extension without RNase T1 digestion. Lane 2–4, structural probing with RNase T1 in the absence of Hfq, in the presence of Hfqwt and in the presence of HfqG29A, respectively. U, A, C, G, sequencing ladder.
![Figure 4. Structural probing of the translation initiation region of rpoS mRNA in complex with DsrA in the absence of Hfq and in the presence of Hfqwt or HfqG29A. First, rpoS mRNA was annealed to DsrA and to the [32P]-5′-end labeled primer. The annealing mix was cooled to 24 °C and divided into three parts followed by incubation for 10 min in the absence and presence of Hfqwt and HfqG29A, respectively. Then RNase T1 was added for 10 min followed by primer extension with AMV reverse transcriptase at 45 °C. The primer extension products were separated on a 8% polyacrylamide-8M urea gel and visualized by a PhosphorImager (Molecular Dynamics). The sequence of the immediate coding region is shown at the left. The start codon is marked by dots. RNase T1 cleavage 3′ of G residues and stop signals of the reverse transcriptase at A residues are indicated at the right. The numbering is given with regard to the A of the start codon (+1). Lane 1, primer extension without RNase T1 digestion. Lane 2–4, structural probing with RNase T1 in the absence of Hfq, in the presence of Hfqwt and in the presence of HfqG29A, respectively. U, A, C, G, sequencing ladder.](/cms/asset/c42b5d08-f412-47ba-b4ea-626bcfaa87bc/krnb_a_10927100_f0004.gif)
CsdA does not rescue the deficiency of HfqG29A
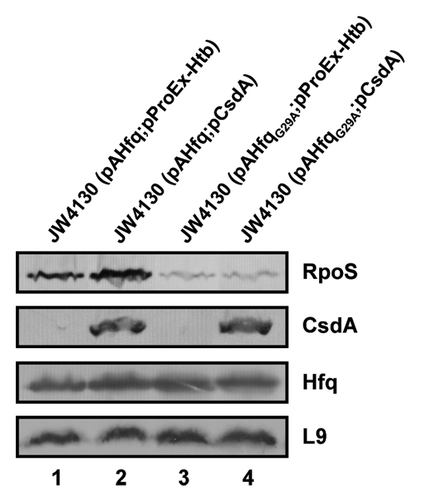
We have previously shown that the CsdA helicase is required for in vivo DsrA·rpoS duplex formation at 24 °C, and have hypothesized that opening of the iss in rpoS by CsdA at low temperature is required for DsrA·rpoS duplex formation.Citation20 We next asked whether overexpression of csdA could also rescue the defect of HfqG29A in translation initiation of rpoS at low temperature. A plasmid borne copy of csdA was co-expressed in strain JW4130 in the presence of HfqG29A and of Hfqwt, respectively. The cells were grown at 24 °C to an OD600 of 0.4 when csdA expression was co-induced with the respective hfq gene/allele. Immunodetection of RpoS revealed no increase of translation in the presence of HfqG29A, whereas in the presence of Hfqwt, synthesis of RpoS slightly increased (). Thus, while CsdA is required for DsrA·rpoS duplex formation,Citation20 it apparently cannot rescue the defect of HfqG29A in terms of converting rpoS mRNA into a translationally compentent form, which assigns distinct roles to CsdA and Hfq toward rpoS translation.
Figure 5. Overexpression of csdA does not rescue rpoS translation in the presence of HfqG29A. Immunodetection of RpoS, CsdA, Hfq, and ribosomal protein L9 in strains JW4130(pAHfq;pProEx-Htb) (lane 1), JW4130(pAHfq;pCsdA) (lane 2), JW4130(pAHfqG29A; pProEx-Htb) (lane 3), and JW4130(pAHfqG29A;pCsdA) (lane 4), respectively. The proteins were visualized as described in the legend to . Only the relevant sections of the immunoblots are shown.
Conclusions
In the context of all work performed on the DsrA/rpoS system, these studies add another facet to Hfq function in translational activation of rpoS mRNA. They suggest that DsrA·rpoS duplex formation, albeit clearly stimulating, is not the ultimate event that results in efficient translation initiation of rpoS mRNA. Besides its function in stabilization of DsrA,Citation4,Citation18 and in accelerating annealing between DsrA and rpoS,Citation23,Citation25 Hfq is apparently also required to restructure the rbs of rpoS mRNA before or upon DsrA·rpoS duplex formation to permit efficient ribosome loading (). Soper et al.Citation23 have put forward a model wherein Hfq remains bound to the rpoS leader after DsrA·rpoS annealing. In light of our results, it seems feasible that lingering of Hfq at the A-rich sites in the rpoS leader brings about efficient ribosome loading on rpoS mRNA by inducing these structural changes. Although some studies indicated an association of Hfq with the translational machinery,Citation34,Citation35 recent studies failed to reveal an interaction of Hfq with ribosomes or with ribosomal protein S1,Citation36 the latter of which is required for translation initiation of structured mRNAs.Citation37,Citation38 Although other interaction partners of Hfq can at present not be excluded, the RNA chaperone function of Hfq might suffice to present the mRNA in a conformation readily accessible for the 30S ribosome.
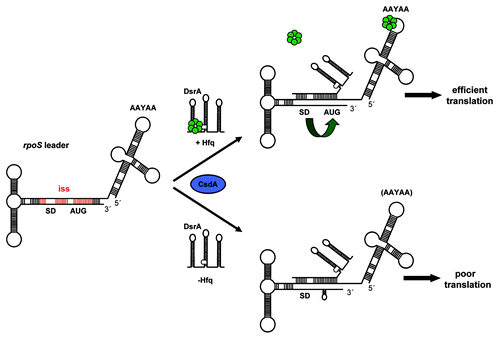
Figure 6. Working model for translational activation of rpoS mRNA at low temperature. The rbs of rpoS mRNA is masked by an iss (in red). In the presence of Hfq (upper scheme) the sRNA DsrA is bound and stabilized by Hfq.Citation4,Citation18,Citation24 CsdA (blue oval) is required for DsrA·rpoS duplex formation,Citation20 which leads to opening of the iss and in displacement of Hfq from DsrA.Citation26 Hfq bound to A-rich segments in the rpoS leader (AAYAA) promotes DsrA·rpoS annealingCitation24 and restructuring of rpoS mRNA into a translationally competent conformer (green bent arrow), which, in turn, permits efficient rpoS translation. In the absence of Hfq (lower scheme), DsrA·rpoS duplex formation occurs when dsrA is overexpressed. However, in the absence of Hfq, rpoS is poorly translated because the local secondary structure within the immediate coding region is not efficiently resolved. The Shine-Dalgarno (SD) sequence and start codon (AUG) in rpoS mRNA are highlighted, Hfq is shown in green.
Materials and Methods
Bacterial strains and plasmids
The E. coli hfq- strain JW4130 has been described.Citation39 The cells were grown in Luria-Bertani (LB) mediumCitation40 at 24 °C supplemented with ampicillin (100 µg/ml), kanamycin (25 µg/ml), or chloramphenicol (20 µg/ml) where appropriate to maintain plasmids.
The plasmid pAHfqCitation13 encoding Hfqwt protein, plasmids pNM12 (empty vector) and pNM13Citation28 used for dsrA overexpression, and plasmids pProEx-Htb (empty vector; Invitrogen) and pCsdACitation41 for overexpression of csdA have been described.
Construction of pAHfqG29A
To introduce the G29A mutation into the hfq sequence, a DNA fragment was generated by means of PCR using the hfq forward primer (5′-GCTCTAGAAA TATAATAGTT TAACTTTAAG AAGGAGATAT ACATATGGCT AAGGGGCAAT CTTTACAAGA TCCGTTCCT-3′), containing an XbaI site (italics), and the reverse primer (5′-AGACTCGATT TGCCCTTGCA GCTTAATAGC ATTCACCAAA-3′). The DNA fragment was used in a second PCR as forward primer together with the hfq reverse primer (5′-GGAATTCCCG TGTAAAAAAA CAGCCCGAAA C-3′), containing an EcoRI site (italics), to generate the full hfqG29A gene. The PCR product was cleaved with XbaI and EcoRI and ligated into the corresponding sites of plasmid pUC19 (New England Biolabs). From the resulting plasmid pUHfqG29A, the hfqG29A allele was re-isolated as a PvuII fragment, and ligated into the EcoRV/NruI sites of plasmid pACYC184 (New England Biolabs), yielding plasmid pAHfqG29A.
Western blot analyses
The cellular levels of Hfqwt, HfqG29A, L9/L14 ribosomal protein(s), RpoS, and CsdA were determined by quantitative immunoblotting. For the experiments shown in and , the E. coli strains JW4130(pACYC184), JW4130(pAHfq), and JW4130(pAHfqG29A) were grown at 24 °C or 37 °C in LB medium supplemented with appropriate antibiotics to maintain plasmids. For the experiment shown in , the E. coli strains JW4130(pNM12), JW4130(pNM13), JW4130(pACYC184), and JW4130(pAHfq) were grown at 24 °C in LB medium supplemented with 0.4% arabinose to induce expression of dsrA from the arabinose promoter of plasmid pNM13. All cells were grown to an OD600 of ~0.4 when equal amounts of cells were withdrawn and boiled in protein sample buffer. For csdA overexpression (), the E. coli hfq- strain JW4130-bearing plasmid pAHfq or pAHfqG29A was co-transformed with either the control plasmid pProEx-HTb (Invitrogen) or with plasmid pCsdA. The strains were grown in LB medium at 24 °C to an OD600 of 0.4 when expression of csdA was induced by addition of 1 mM IPTG (final concentration). After 1 h of growth equal amounts of the cells were withdrawn and boiled in protein sample buffer. Equal amounts of total protein were separated on 12% SDS–polyacrylamide gels and blotted onto a nitrocellulose membrane (GE-healthcare). The blots were blocked with 5% dry milk in TBS buffer, and then probed with anti-Hfq (lab. stock) anti-L9/-L14 (lab. stock), anti-His-tag (Sigma), or anti-RpoS (Santa Cruz Biotech) antibodies. The antibody–antigen complexes were visualized with alkaline–phosphatase conjugated secondary antibodies (Sigma) using the chromogenic substrates nitro blue tetrazolium chloride (NBT) and 5-Bromo-4-chloro-3-indolyl phosphate (BCIP).
Northern blot analyses
The steady-state levels of DsrA were determined by northern blot analysis using 10 µg of total RNA. RNA extraction was performed by the Trizol method (Ambion). The RNA samples were denatured for 5 min at 85 °C in RNA loading dye, separated on 8% polyacrylamide-8 M urea gels, and then transferred to a nylon membrane (Amersham Hybond-N) by electroblotting. The RNA was cross-linked to the membrane by exposure to UV light. The membrane was hybridized with DsrA-specific [γ-32P]-5′-end-labeled (Amersham Pharmacia Biotech) oligonucleotide (5′-TCGTTACACC AGGAAATCTG ATGT-3′) or 5S-rRNA-specific [32P]-5′-end labeled oligonucleotide (5′-GGTGGGACCA CCGCGCTACG GCCGCCAGGC-3′). Hybridization signals were visualized using a PhosphorImager (Molecular Dynamics).
Primer extension assays
For the experiment shown in , the E. coli strains JW4130(pNM12), JW4130(pNM13), JW4130(pACYC184),and JW4130(pAHfq) were grown at 24 °C in LB medium supplemented with 0.4% arabinose to induce expression of dsrA from the arabinose promoter of plasmid pNM13. For the experiment shown in , the E. coli hfq- strain JW4130 bearing plasmids pACYC184, pAHfqG29A, and pAHfq, respectively, was grown at 24 °C. At an OD600 of 0.4, samples for RNA isolation and for western blot analysis were withdrawn. Isolation of total RNA was performed using the Trizol method (Ambion). The primer extension analysis, used to detect the RNase III-specific cleavage at position G−112/A−109 in rpoS mRNA, was performed as described.Citation21 In brief, two units of AMV reverse transcriptase (Promega) were used for 20 µg of purified total RNA primed with the rpoS-specific [32P]-5′-end labeled oligonucleotide (5′-TCCGTTCTCA TCAAATTCCG CATC-3′). The extension products along with a sequencing ladder, which was prepared using the 5′-segment (nt -564 to +188) of rpoS mRNA as a template, were resolved on a 8% polyacrylamide-8M urea gel. The resulting signals were visualized by a PhosphorImager (Molecular Dynamics).
RNA preparation for in vitro studies
To prepare full-length rpoS mRNA, plasmid pUrpoS16Citation13 digested with EcoRI was used as template for in vitro transcription with T7 RNA polymerase (Promega). Preparation of DsrA RNA was performed as described.Citation21 The run-off transcripts were purified on 8% polyacrylamide-8M urea gels following standard procedures. The RNA concentration was determined by measuring the A260.
RNaseT1 probing
Hfqwt and HfqG29A proteins were purified from strain AM111F’Citation42 harboring plasmids pUH5Citation43 (Hfqwt) and pUHfqG29A (HfqG29A), respectively, as described.Citation44 1.8 pmol of full-length rpoS mRNA, 3.6 pmol of DsrA RNA, and 2 pmol [32P]-5′-end labeled primer (5′-TTTTCGTCAA AAACCTCAAC TCCG-3′; corresponds to nucleotides +59 to +83 in the rpoS gene) were annealed by incubation at 85 °C for 3 min in ddH20. Then, the complexes were cooled to 24 °C and concentrated AMV buffer (Promega) was added. The annealing mix was divided into three parts. One aliquot was further incubated in the absence of Hfq, whereas the other two aliquots were incubated in the presence of 5 pmol Hfqwt and 5 pmol HfqG29A, respectively for 10 min at 24 °C. Then, 2 U of RNase T1 dilution (Ambion) was added to the reactions. The samples were further incubated for 5 min at 24 °C, before they were heated to 45 °C. Then, dNTPs and reverse transcriptase (2 U AMV) were added. The primer extension products along with a sequencing ladder, which was prepared using the 5′-segment (nt -564 to +188) of rpoS mRNA as a template, were resolved on 8% polyacrylamide-8M urea gels. The resulting signals were visualized by a PhosphorImager (Molecular Dynamics).
Additional material
Download Zip (475.1 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The work was supported by the Austrian Science Fund through the Special Research Program RNA-REG F43, subproject AF4311 (UB), and the doctoral program RNA-Biology W-1207 (HH). We thank Drs S Gottesman and T Bizebard for providing materials.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/rnabiology/article/27100/
References
- Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell 2009; 101:117 - 31; http://dx.doi.org/10.1042/BC20070137; PMID: 19076068
- Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev 2010; 34:866 - 82; PMID: 20662934
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol 2011; 9:578 - 89; http://dx.doi.org/10.1038/nrmicro2615; PMID: 21760622
- Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003; 9:1308 - 14; http://dx.doi.org/10.1261/rna.5850703; PMID: 14561880
- Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli.. Genes Dev 2003; 17:2374 - 83; http://dx.doi.org/10.1101/gad.1127103; PMID: 12975324
- Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J 2002; 21:3546 - 56; http://dx.doi.org/10.1093/emboj/cdf322; PMID: 12093755
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 2004; 11:1206 - 14; http://dx.doi.org/10.1038/nsmb858; PMID: 15531892
- Ribeiro EdeA Jr., Beich-Frandsen M, Konarev PV, Shang W, Večerek B, Kontaxis G, Hämmerle H, Peterlik H, Svergun DI, Bläsi U, et al. Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res 2012; 40:8072 - 84; http://dx.doi.org/10.1093/nar/gks510; PMID: 22718981
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 2013; 425:3678 - 97; http://dx.doi.org/10.1016/j.jmb.2013.01.006
- Vincent HA, Henderson CA, Stone CM, Cary PD, Gowers DM, Sobott F, Taylor JE, Callaghan AJ. The low-resolution solution structure of Vibrio cholerae Hfq in complex with Qrr1 sRNA. Nucleic Acids Res 2012; 40:8698 - 710; http://dx.doi.org/10.1093/nar/gks582; PMID: 22730296
- Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A 2009; 106:19292 - 7; http://dx.doi.org/10.1073/pnas.0908744106; PMID: 19889981
- Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci U S A 2012; 109:9396 - 401; http://dx.doi.org/10.1073/pnas.1202521109; PMID: 22645344
- Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res 2008; 36:133 - 43; http://dx.doi.org/10.1093/nar/gkm985; PMID: 18000007
- Moll I, Leitsch D, Steinhauser T, Bläsi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep 2003; 4:284 - 9; http://dx.doi.org/10.1038/sj.embor.embor772; PMID: 12634847
- Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J 2004; 23:396 - 405; http://dx.doi.org/10.1038/sj.emboj.7600058; PMID: 14739933
- Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 1996; 15:3993 - 4000; PMID: 8670904
- Repoila F, Gottesman S. Temperature sensing by the dsrA promoter. J Bacteriol 2003; 185:6609 - 14; http://dx.doi.org/10.1128/JB.185.22.6609-6614.2003; PMID: 14594834
- Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol 2001; 183:1997 - 2005; http://dx.doi.org/10.1128/JB.183.6.1997-2005.2001; PMID: 11222598
- Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci U S A 1998; 95:12456 - 61; http://dx.doi.org/10.1073/pnas.95.21.12456; PMID: 9770507
- Resch A, Večerek B, Palavra K, Bläsi U. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol 2010; 7:796 - 802; http://dx.doi.org/10.4161/rna.7.6.13768; PMID: 21045550
- Resch A, Afonyushkin T, Lombo TB, McDowall KJ, Bläsi U, Kaberdin VR. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 2008; 14:454 - 9; http://dx.doi.org/10.1261/rna.603108; PMID: 18192613
- Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA 2011; 17:489 - 500; http://dx.doi.org/10.1261/rna.2452111; PMID: 21205841
- Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 2008; 14:1907 - 17; http://dx.doi.org/10.1261/rna.1110608; PMID: 18658123
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A 2010; 107:9602 - 7; http://dx.doi.org/10.1073/pnas.1004435107; PMID: 20457943
- Hopkins JF, Panja S, Woodson SA. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res 2011; 39:5193 - 202; http://dx.doi.org/10.1093/nar/gkr062; PMID: 21378124
- Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol 2004; 344:1211 - 23; http://dx.doi.org/10.1016/j.jmb.2004.10.006; PMID: 15561140
- Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA 2003; 9:33 - 43; http://dx.doi.org/10.1261/rna.2570803; PMID: 12554874
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A 1998; 95:12462 - 7; http://dx.doi.org/10.1073/pnas.95.21.12462; PMID: 9770508
- Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA 2011; 17:1544 - 50; http://dx.doi.org/10.1261/rna.2767211; PMID: 21705431
- Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol 2001; 39:1382 - 94; http://dx.doi.org/10.1111/j.1365-2958.2001.02329.x; PMID: 11251852
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 2010; 29:3094 - 107; http://dx.doi.org/10.1038/emboj.2010.179; PMID: 20683441
- McCullen CA, Benhammou JN, Majdalani N, Gottesman S. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol 2010; 192:5559 - 71; http://dx.doi.org/10.1128/JB.00464-10; PMID: 20802038
- Hämmerle H, Beich-Frandsen M, Večerek B, Rajkowitsch L, Carugo O, Djinović-Carugo K, Bläsi U. Structural and biochemical studies on ATP binding and hydrolysis by the Escherichia coli RNA chaperone Hfq. PLoS One 2012; 7:e50892; http://dx.doi.org/10.1371/journal.pone.0050892; PMID: 23226421
- Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J Bacteriol 1994; 176:531 - 4; PMID: 8288550
- Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry 2003; 42:8022 - 34; http://dx.doi.org/10.1021/bi020638i; PMID: 12834354
- Večerek B, Beich-Frandsen M, Resch A, Bläsi U. Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res 2010; 38:1284 - 93; http://dx.doi.org/10.1093/nar/gkp1125; PMID: 19969548
- Tedin K, Resch A, Bläsi U. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′ leader sequence. Mol Microbiol 1997; 25:189 - 99; http://dx.doi.org/10.1046/j.1365-2958.1997.4421810.x; PMID: 11902720
- Sørensen MA, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo.. J Mol Biol 1998; 280:561 - 9; http://dx.doi.org/10.1006/jmbi.1998.1909; PMID: 9677288
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006; 2:0008; http://dx.doi.org/10.1038/msb4100050; PMID: 16738554
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory 1972; Cold Spring Harbor.
- Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 2004; 43:7857 - 66; http://dx.doi.org/10.1021/bi049852s; PMID: 15196029
- Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli.. Genes Dev 1996; 10:1143 - 51; http://dx.doi.org/10.1101/gad.10.9.1143; PMID: 8654929
- Večerek B, Moll I, Afonyushkin T, Kaberdin V, Bläsi U. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli.. Mol Microbiol 2003; 50:897 - 909; http://dx.doi.org/10.1046/j.1365-2958.2003.03727.x; PMID: 14617150
- Beich-Frandsen M, Večerek B, Konarev PV, Sjöblom B, Kloiber K, Hämmerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA, et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res 2011; 39:4900 - 15; http://dx.doi.org/10.1093/nar/gkq1346; PMID: 21330354