Abstract
Tight junctions are the critical intercellular structure required to establish an epithelial barrier. Among the several classes of proteins required to form tight junctions are the tetraspan transmembrane proteins known as claudins that directly determine paracellular permeability. Considerable progress has been made in understanding how incorporation of different claudins into tight junctions increase or decrease paracellular permeability and ion selectivity. However, it has proven difficult to identify discrete steps in claudin assembly and whether claudins exist in distinct oligomerization states prior to tight junction assembly. Studies of homomeric and heteromeric claudin-claudin interactions using complementary techniques suggest a diversity of pathways used by different claudins to oligomerize and integrate into tight junctions.
Introduction
Tight junctions regulate the fluid flow between cells across the apical / basolateral axis. Several classes of proteins are required to assemble tight junctions, including single and multi-pass transmembrane proteins, cytosolic scaffold proteins and cytoskeletal associated proteins.Citation1 These proteins all coordinate to produce a functional paracellular barrier, however, it is claudin family proteins that are central to the regulation of tight junctional permeability.Citation2-Citation5 This is due to two functional classes of claudins, pore forming claudins which create paracellular ion channels and sealing claudins which more generally restrict paracellular flux.
Although there is considerable literature demonstrating specific roles for different claudins in regulating epithelial and endothelial barrier function, how these proteins are specifically assembled into tight junction strands is less well understood. The scope of this article is to summarize current progress in understanding how claudins become integrated into tight junction strands with particular emphasis on what is known about stable higher order claudin intermediates which form prior to incorporation into tight junctions.
Claudins and Tetraspanins
Claudins have four transmembrane domains with the N- and C-terminus oriented toward the cytoplasm (). The N-terminus is short containing roughly 7 amino acids and is readily modified with epitope tags or fluorescent proteins which do not interfere with their assembly or function. The C-terminus is considerably more variable, ranging from 20 to over 60 amino acids. The extreme C-terminus of virtually all claudins has the capacity to bind to the cytosolic scaffold proteins zonula occludens-1 (ZO-1) and ZO-2 via a Post-synaptic density 95, Discs large, ZO-1 (PDZ) binding motif.Citation6 ZO-1 and/or ZO-2 binding promotes tight junction assembly by tethering claudins to the actin cytoskeleton.Citation7 However, other classes of scaffold proteins, such as PALS-1-associated tight junction protein (PATJ), have also been found to bind to claudins which further regulate junction assembly.Citation8,Citation9
Figure 1. Claudin secondary structure. (A) Claudin line diagram showing key features including the two extracellular loop (EL) domains, where EL1 contains a putative di-sulfide bond (S-S). Cylinders represent predicted transmembrane α-helical domains. Also highlighted are two palmitoylation motifs (“P”) and the PDZ binding motif at the extreme C-terminus of the protein. (B) Claudin conformation in the plane of the membrane, showing the four transmembrane α-helical domains as a tightly packed complex. Adapted from.Citation4
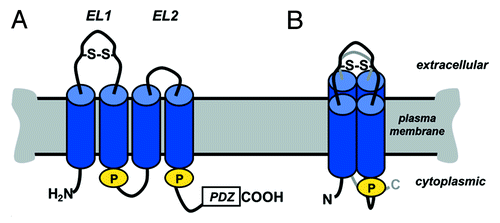
Claudins contain two extracellular loop domains which mediate intercellular interactions and are required for control of paracellular permeability.Citation10,Citation11 The first extracellular loop domain contains cysteines which likely stabilize its secondary structure, since mutating either or both of these cysteines disrupts the ability of cldn-5 to form functional tight junctions.Citation11 The second extracellular loop is smaller (~16 amino acids) and has been modeled to form a helix-turn-helix structure.Citation12 Claudin selectivity is mainly controlled by motifs in the first extracellular loop domain, however, claudin-claudin interactions are determined by both extracellular loop domains.Citation5,Citation13
Claudins are tetraspan proteins, however, they are not directly homologous with the proteins directly classified as tetraspanins (e.g., CD9, CD81, CD151).Citation14,Citation15 In particular, claudins have considerably more protein extending outside either the extracellular or cytoplasmic faces of the membrane as opposed to tetraspanins which are almost entirely membrane embedded.Citation16 However, claudins and tetraspanins both preferentially partition into cholesterol- and sphingolipid-enriched membrane microdomains (also referred to as lipid rafts), suggesting that they interact with membrane lipids in a comparable manner.Citation14,Citation15,Citation17-Citation19 This is due in part to the fact that both claudins and tetraspanins are palmitoylated, which promotes their partitioning into membrane microdomains and assembly into higher order structures.Citation14,Citation15,Citation19
Tetraspanins have been found to promote the trafficking and assembly of several classes of proteins, most notably integrins and MHC class II proteins.Citation14,Citation15 Tetraspanin-directed membrane assembly is complex, since these proteins interact weakly with each other and with other substrates. Also, tetraspanin-protein interactions are initiated in several distinct intracellular organelles, including the endoplasmic reticulum (ER) and Golgi apparatus. Tetraspanins have also been associated with large-scale heterogeneous membrane lipid–protein complexes,Citation14 which are less tractable to biochemical analysis than more classical membrane microdomains.Citation20-Citation22
Given roles for tetraspanins in organizing membrane structure, it seems likely that these proteins will participate in tight junction formation as well. Consistent with this hypothesis, proteomic analysis of isolated liver junctions demonstrates that they are enriched for claudin-1, claudin-3 and several tetraspanins including CD81.Citation23 Consistent with a more direct interaction, claudin-1 associates with the tetraspanin CD81 as part of the hepatitis C virus receptor complex.Citation24 Moreover, claudin-1 and CD9 partition into comparable membrane microdomains and can be chemically crosslinked, suggesting a direct interaction.Citation25 Other claudins have been found to weakly associate with CD9.Citation25
Another pathway linking claudins to tetraspanins is via the protein Epithelial Cell Adhesion Molecule (EpCAM). EpCAM has several functions and has the capacity to promote cell adhesion, however it does not show homology to Ig-family CAMs and instead has an extracellular domain more closely related to nidogens.Citation26 There are several lines of evidence that claudin-7 interacts directly with EpCAM and forms a complex with the tetraspanins CD9, CD44v6 and CO-029.Citation27,Citation28 This complex can also include some claudins (e.g., claudin-1) but not others (e.g., claudin-2, claudin-4) suggesting specific heteromeric interactions with claudin-7 (see below).Citation29
EpCAM and claudin-7 tend to be overexpressed by some highly metastatic tumors,Citation28,Citation30,Citation31 however, EpCAM also has a role in regulating tight junction assembly, since EpCAM deficient mice have decreased incorporation of several claudins into junctions.Citation32 In fact, EpCAM depleted intestinal epithelial cells show slower rates of barrier formation than cells with normal EpCAM expression.Citation29 Interestingly, claudin-7/EpCAM complexes tend to be more closely associated with non junctional lateral aspects of epithelial cells although there is some EpCAM associated with tight junctions as well.Citation29,Citation32 This non-junctional pool of claudin-7 requires EpCAM and has an as yet unknown function. These links between tetraspanins and claudins are intriguing and in need of further study, particularly given evidence that there are distinct pools of membrane microdomains enriched for different tetraspanins that regulate claudin trafficking and assembly.
Heteromeric Claudin Interactions
Claudins on adjacent cells can bind to each other via the extracellular domains across tight junctions, via trans interactions (). Virtually all claudins have been found to homotypically interact.Citation33 Heterotypic trans interactions have been most conclusively demonstrated to involve claudin-3, which binds to claudin-1, claudin-2 and claudin-5 as well as homotypically to claudin-3 (33–35). There is also some evidence that claudin-1 and claudin-5 may heterotypically interact,Citation36 although this expected to be weak compared with claudin-3/claudin-5 trans binding.Citation33 On the other hand, there is ample evidence that several claudins are heterotypically incompatible.Citation37-Citation39
Figure 2. Pathways for claudin oligomerization. In this diagram, the early pathway depicts claudin oligomerization (cis interactions) occurring in the endoplasmic reticulum (ER), the late pathway shows oligomerization at the trans Golgi network (TGN). These represent two extremes since oligomerization could also occur in other compartments, such as the ER-Golgi intermediate compartment (ERGIC) or the Golgi apparatus itself. Alternatively, oligomerization could be driven by processes associated with tight junction assembly, including scaffold protein/cytoskeletal tethering and trans interactions between claudins on adjacent cells. For simplicity, scaffold proteins are represented by ZO-1, other scaffold proteins also contribute to stabilize claudins by linking them to the cytoskeleton. Also, this diagram only shows homomeric/homotypic interactions, heteromeric and heterotypic claudin-claudin interactions or between claudins and Marvel proteins are expected to follow comparable pathways.
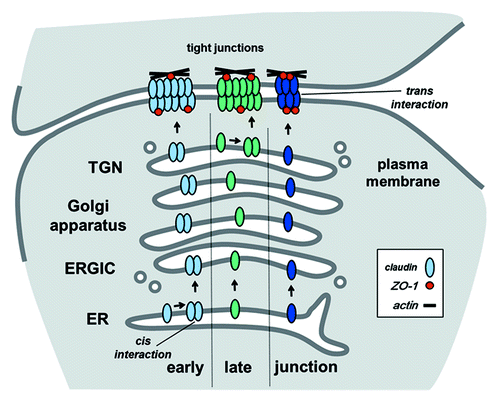
Claudins expressed by the same cell also have the capacity to interact in the plane of the membrane via cis interactions, which is also known as oligomerization (). Heteromeric claudin oligomerization is much more frequent than heterotypic binding. Moreover, not all claudins form homomeric oligomers (). Heteromer specificity has the capacity to create subdomains enriched for specific claudins which, in turn, have the potential to modulate tight junction permeability. Tight junction subdomains can be further stabilized by homotypic interactions across junctions, e.g., through the formation of homotypic heteromers. As a specific example, cells in the reticular lamina of the inner ear have two classes of junctional subdomains, one class enriched for heteromeric claudin-6 and claudin-9 and another enriched for claudin-14, which serve distinct functions.Citation39 Although tight junctions do not usually have such extreme differences in claudin localization, it is likely that most have fine structure driven by claudin heterogeneity which helps regulate barrier function.
Table 1. Homomeric and heteromeric claudin-claudin interactions
The coordinate interaction between claudin-16 and claudin-19 provides another example of heteromer driven functionality, since mutations in either of these two claudins cause the renal disease Familial Hypomagnesemia with Hypercalciuria and NephroCalcinosis (FHHNC).Citation40,Citation41 Claudin-16 and claudin-19 hetero-oligomerize to form sodium-selective tight junction pores which provides an indirect mechanism for regulation of magnesium resorption.Citation42,Citation43 Critically, transport of claudin-16 and claudin-19 are mutually dependent. In the absence of claudin-16, claudin-19 is retained in the perinuclear region of the cell, suggesting Golgi apparatus retention, although this region of the cell may also reflect endosome localization.Citation44,Citation45 Coordinate trafficking of claudin-16 and claudin-19 is also observed in the intact kidney, indicating that these two claudins form obligate heteromers in order to function.Citation42 Interestingly, claudin-19 can also hetero-oligomerize with claudin-10a and claudin-18, however, claudin-16 does not.Citation42 Since claudin-10a and claudin-18 form tight junctions in FHHNC, co-chaperone function of claudin-19 is not required for these claudins to integrate into tight junctions. In addition, claudin-18 forms functional tight junctions in the lung and stomach, which do not express claudin-19.Citation46 Obligate heteromer formation has also been implicated for trafficking of claudin-4 and claudin-8 in the kidney.Citation47 However, claudin-4 properly traffics and forms tight junctions in transfected claudin null HeLa cells,Citation33 suggesting that this may be a unique feature of claudin-4 transport in the kidney. For instance, renal epithelial cells may lack chaperones present in other cell types that are compensated for by claudin-8. Considering that claudin-4 expression is much more prevalent than claudin-8, multiple mechanisms to regulate claudin-4 assembly into tight junctions are likely.
Other examples of claudin heteromers are shown in . Two in situ techniques have been primarily used to measure heteromeric interactions: Fluorescence Resonance Energy Transfer (FRET) and yeast two hybrid assays. FRET has been used to study both homomeric and heteromeric interactions, with particular emphasis on claudins expressed by endothelial cells.Citation12,Citation36 C-terminal Cyan Fluorescent Protein (CFP) or Yellow Fluorescent Protein (YFP) tagged claudins used as donor and acceptor fluorophore block binding to ZO-1 and ZO-2, which helps ensure that a direct heteromer interaction is being measured. Also, subconfluent cells or claudin point mutants incapable of trans binding enable heteromeric interactions to be distinguished from heterotypic interactions. Using this approach, Piontek and coworkersCitation36 have found that FRET efficiency differed for different pairs of claudins, ranging from claudin-5 homomers which had a high FRET signal to claudin-3 / claudin-5 heteromers which had lower, but detectable FRET. Other pairs, such as claudin-2 and claudin-3 showed no detectable interaction. Moreover, claudin-3 / claudin-5 FRET was enhanced as a result of tight junction formation. These data support the model where homotypic heteromers are a major component of fully formed tight junctions.
Hou et al. pioneered the use of a yeast two hybrid assay to assess homomer and heteromer compatibility since yeast do not form tight junctions and thus do not allow heterotypic interactions between transfected mammalian claudins.Citation42,Citation44,Citation47 This yeast two-hybrid assay was used to show that claudin-16 and claudin-19 hetero-oligomerize.Citation42 Interestingly, several classes of claudins have been found to be heteromerically incompatible by yeast two hybrid analysis (). These findings are not always concordant with results obtained by mammalian expression systems. For instance, claudin-3 homomers were detected by FRET,Citation36 yet claudin-3 did not produce a detectable homo-oligmerization signature by yeast two hybrid analysis.Citation47 The mechanistic explanation for this difference is not known at present. Since membrane microdomains are critical for claudin assembly into tight junctions, one intriguing possibility is that claudin-claudin interactions are sensitive to membrane microenvironment which significantly differs between yeast and mammalian cells.Citation48,Citation49 In this case, a lack of cis interaction in yeasts may reflect claudin specific differences in membrane partitioning and/or conformation. If this is the case, perturbation of lipid composition is likely to have differential effects on different claudins. Consistent with this possibility, cholesterol depletion of Caco2 cells using methyl β-cyclodextrin preferentially inhibited partitioning of claudin-3 and claudin-4 into detergent resistant membrane microdomains as opposed to claudin-1 which was resistant to methyl β-cyclodextrin.Citation18
Interactions of Claudins with Marvel Proteins
The Marvel proteins occludin, tricellulin and MarvelD3 represent the other major class of tetraspan proteins associated with tight junctions.Citation50-Citation52 Several lines of evidence support interactions between occludin and claudins. For example, occludin co-purifies with claudins directly binding to Clostridium Perfringens Enterotoxin (CPE).Citation53,Citation54 Since CPE cannot directly bind occludin,Citation55,Citation56 a claudin-occludin interaction is required for incorporation of occludin in the complex.Citation57 Occludin also directly binds to claudin-1 extracellular loop mimetic peptides, suggesting the potential for heteromeric and/or heterotypic interactions between occludin and claudins.Citation58 In addition, occludin is present in a discrete high molecular weight (mega Dalton) complex isolated from dodecylmaltoside solubilized liver which also contains claudin-1 and claudin-2.Citation59
FRET based assays have also been used to examine cis interactions between occludin and different claudins in binary mixtures using transfected HEK cells.Citation60 In this system, occludin interacts with several claudins to varying degrees where occludin and claudin-1 showed a strong cis interaction with claudin-1 and a weaker interaction with claudin-2 and claudin-4. Claudin-3, claudin-5 and claudin-11 interacted poorly or not at all with occludin. By contrast, Marvel D3 interacts with claudin-3 to a level comparable with claudin-4.Citation60 Both tricellulin and Marvel D3 interact with claudin-5 and not with claudin-2 or claudin-11. Interestingly, in a trinary transfected HEK cell system, occludin effectively competes for tricellulin in interacting with claudin-1 which may play a role in directing tricellulin to tricellular contacts.Citation61,Citation62 Claudin expression also promoted the localization of Marvel proteins to intercellular contacts, consistent with a dual feedback mechanism where these proteins co-assemble into tight junctions.
Taken together these results suggest a role for Marvel proteins in organizing the fine structure of tight junction claudin composition. However the relative stability of these interactions remains to be determined. In particular, studies using Fluorescence Recovery After Photobleaching (FRAP) have shown that claudins tend to be stable components of tight junctions, whereas occludin is significantly more mobile.Citation63 Thus, occludin / claudin interactions are likely to be more transient than claudin / claudin interactions, particularly in a fully assembled tight junction. This leads to a model where occludin and other Marvel proteins may act as co-chaperones to promote claudin transport through the cell and vice versa which then changes once the claudins are assembled into fully formed tight junctions.
Size of Claudin Oligomers
Biochemical analysis of solubilized claudins suggests that claudins form stable intermediates. Claudin complexes as large as hexamers have been detected by native gel electrophoresis of samples solubilized by perfluorooctanoic acid (PFO).Citation35,Citation64 Size analysis of solubilized tight junction protein complexes is clearly informative. However, using this approach to analyze claudin oligomerization is confounded by the potential for heterotypic binding and/or stabilization by a shared interaction with another protein, such as occludin or ZO-1. These factors can be partially controlled by using transfected claudin null cell lines, such as fibroblasts, HEK and HeLa cells.Citation12,Citation33-Citation35
Critically, oligomer size differs for different claudins and solubilization conditions. For instance, claudin-4 expressed by transfected Sf9 insect cells resolved as stable hexamers when solubilized in PFO,Citation64 whereas PFO-solubilized claudins from transfected NIH 3T3 cells resolved as either monomers (in the cases of claudin-1 and claudin-4) or trimers/tetramers (for claudin-5).Citation35 For claudin-1 and claudin-5, 3T3 cells solubilized in 0.1% Triton X-100 and then resolved by native gel electrophoresis produced comparable results. However, claudin-3 expressed by 3T3 cells and solubilized in Triton X-100 appeared to remain as stable tetramers.Citation35 Moreover, dimers of claudin-1, claudin-2 and claudin-7 were stable in dodecylmaltoside treated MDCK cells, whereas claudin-4 was completely solubilized under comparable conditions.Citation59 In this study, claudin-2 was demonstrated to form homodimers, whether the other claudins were homo or heterodimers has not been determined.
Sites of Claudin Oligomerization
represents three potential models for claudin oligomerization, early oligomerization, late oligomerization or oligomerization driven following assembly into tight junctions. The early pathway corresponds to the classical ER quality control pathway for multimeric transmembrane protein oligomerization.Citation65,Citation66 Consistent with a requirement for claudin quality control early in the secretory pathway, mutant claudins which are misfolded accumulate in the ER, although misfolded claudins more typically are retained in the perinuclear region of the cell, e.g., the Golgi apparatus.Citation12,Citation45
Does retention of misfolded claudins reflect control of oligomerization in the ER? This is not likely to be the case. For example, a mutant form of claudin-16 associated with FHHNC (claudin-16-R149L) is retained in the ER but lacks the ability to exert a dominant negative effect on claudin-19 which instead accumulates in the perinuclear region of the cell.Citation44 This is despite the normal co-trafficking of these two claudins in the healthy kidney epithelium. Another line of evidence comes from an analysis of claudins tagged with an ER retention signal, His-Lys-Lys-Ser-Leu (HKKSL).Citation4,Citation67,Citation68 Claudin-3-HKKSL and claudin-4-HKKSL are retained in the ER, however neither of these claudins has a dominant negative effect on untagged claudin-3 or claudin-4.Citation4 By contrast wild type claudin-3 and claudin-4 readily form heteromeric oligomers and tight junctions, indicating that claudin-3 / claudin-4 hetero-oligomerization must occur beyond the ER.Citation33,Citation47
Given the potential for claudin-claudin interactions in the late secretory pathway, it is useful to compare claudins with another class of tetraspan junction proteins, connexins, which form gap junction channels. Interestingly, connexin43 normally oligomerizes in the trans Golgi network (TGN),Citation69,Citation70 corresponding to the late oligomerization pathway shown in . Mechanistically, connexin43 monomers are stabilized through an interaction with an ER chaperone protein ERp29.Citation68 The connexin43/ERp29 complex exits the ER and is transported to the Golgi apparatus where ERp29 dissociates and enables connexin oligomerization to proceed. Although data suggests that claudins do not interact with ERp29,Citation68 there may be other chaperones which perform a comparable function to stabilize monomeric claudins.
However, in contrast to HKKSL-tagged claudins, connexin43-HKKSL shows a clear, specific dominant negative effect on the trafficking of connexin46 to the plasma membrane.Citation4,Citation67,Citation71 In this case, connexin43-HKKSL mimics a frameshift mutant connexin43 associated with oculodentodigital dysplasia (fs260) which is retained in the ER and prevents exit of wild type connexin43 from the ER due to a dominant negative effect.Citation72 Why do HKKSL-tagged and mutant connexin43 act as dominant negatives, while comparable claudins do not? In part, this is due to the fact that the connexin quality control pathway is easily saturated.Citation73 Thus, ER retained connexins act as dominant negatives by overwhelming ER resident chaperones which enables premature oligomerization to occur.
Moreover, it is well established that connexins form stable hexamers which have been structurally resolved at high resolution.Citation74 Thus, the inability of ER retained claudins to act as dominant negatives suggests several possibilities. First, it seems likely that claudin oligomers are relatively unstable in situ; this is underscored by the relative difficulty of biochemically isolating claudin complexes due to their instability under a wide array of detergent solubilization conditions.Citation64 Second, a lack of claudin oligomerization in the ER also indicates that they either do not require chaperones to be stabilized as monomers or do not reach levels of expression that saturate normal ER quality control. Possible control points for claudins which would not be saturable in the ER include a requirement for specific membrane microdomains to support oligomerization or redox control (e.g., regulated formation of extracellular loop disulfide bonds).
Nonetheless, some claudins have been found to stably interact inside the secretory pathway since cells which lack tight junctions or are plated at subconfluent density exhibit formation of claudin oligomers.Citation47,Citation64,Citation75 However, it seems unlikely that this would occur in the ER. Instead, claudin oligomerization is more likely to happen in the TGN or another late secretory pathway. As described above, given the role of claudin partitioning into membrane microdomains in tight junction assembly and that this largely occurs in the Golgi apparatus, claudin oligomerization via the late secretory pathway seems like the most likely possibility for formation of stable claudin-claudin intermediates. Given the structural diversity of different claudin family members, it is unlikely that all claudins will oligomerize via the same pathway. In fact, different connexins oligomerize in different intracellular compartments depending on their structure,Citation68,Citation71,Citation76 suggesting the potential for a similar model where different claudins oligomerize via different intracellular pathways.
Summary and Perspective
Claudins play a unique role in regulating paracellular permeability. In order for claudins to effectively function, they need to be properly synthesized, transported to the cell surface and integrated into tight junctions. This includes a requirement for specific interactions with other claudins and other classes of transmembrane and cytosolic tight junction proteins. Unlike many other classes of junction proteins, claudins have proven difficult to analyze via traditional methods. In particular, they are limited in the ability to form highly stable, well defined assembly intermediates tractable to biochemical analysis. This is likely due to the inherent properties of claudins as highly hydrophobic proteins which form large complexes. Cis interactions within the complex are likely to be low affinity and dependent on local membrane composition, comparable to tetraspanins. However, once assembled into tight junctions, higher affinity trans interactions and scaffold protein binding predominate. Given this, in situ methods have proven useful to identify steps in claudin processing, with the caveat that they sometimes yield conflicting results. Any discordant results most likely reflect the roles that claudin heterogeneity and cell architecture have in regulating how tight junctions form and function and underscore the need for new approaches to study how claudins interact. For example, super resolution fluorescence microscopy was recently used to demonstrate that claudin-3 and claudin-5 assemble into tight junctions that have fundamentally distinct architecture at 20 nm resolution.Citation77 Using this approach applied to double or triple labeled samples would help measure claudin fine structure in intracellular organelles and tight junctions.
Moreover, as new claudin mutations associated with human disease are discovered, it is likely that there will be more instances where there will be an induced dominant negative effect as a result of interference with normal claudin transport and assembly. This could either be due to a lack of co-chaperone activity, as is the case in FHHNC or perhaps due to an induced novel interaction, as is the case in other classes of junction-associated disease. Defining stages of claudin olgomerization will likely provide control points with the potential for pharmacologic manipulation which either strengthen tight junction barriers to prevent tissue injury or to temporarily disrupt barriers as a route to drug delivery.
Acknowledgments
I thank Sam Molina for critical review of the manuscript. This work was supported by National Institutes of Health grants HL-083120, AA-013757 and by the Emory University Research Committee.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2001; 2:285 - 93; http://dx.doi.org/10.1038/35067088; PMID: 11283726
- Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 2009; 1:a002584; http://dx.doi.org/10.1101/cshperspect.a002584; PMID: 20066090
- Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta 2008; 1778:631 - 45; http://dx.doi.org/10.1016/j.bbamem.2007.10.018; PMID: 18036336
- Overgaard CE, Daugherty BL, Mitchell LA, Koval M. Claudins: control of barrier function and regulation in response to oxidant stress. Antioxid Redox Signal 2011; 15:1179 - 93; http://dx.doi.org/10.1089/ars.2011.3893; PMID: 21275791
- Engelund MB, Yu AS, Li J, Madsen SS, Færgeman NJ, Tipsmark CK. Functional characterization and localization of a gill-specific claudin isoform in Atlantic salmon. Am J Physiol Regul Integr Comp Physiol 2012; 302:R300 - 11; http://dx.doi.org/10.1152/ajpregu.00286.2011; PMID: 21975646
- Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol 2006; 68:403 - 29; http://dx.doi.org/10.1146/annurev.physiol.68.040104.131404; PMID: 16460278
- Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol 1999; 147:1351 - 63; http://dx.doi.org/10.1083/jcb.147.6.1351; PMID: 10601346
- Poliak S, Matlis S, Ullmer C, Scherer SS, Peles E. Distinct claudins and associated PDZ proteins form different autotypic tight junctions in myelinating Schwann cells. J Cell Biol 2002; 159:361 - 72; http://dx.doi.org/10.1083/jcb.200207050; PMID: 12403818
- Roh MH, Liu CJ, Laurinec S, Margolis B. The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. J Biol Chem 2002; 277:27501 - 9; http://dx.doi.org/10.1074/jbc.M201177200; PMID: 12021270
- Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol 2002; 283:C142 - 7; PMID: 12055082
- Wen H, Watry DD, Marcondes MC, Fox HS. Selective decrease in paracellular conductance of tight junctions: role of the first extracellular domain of claudin-5. Mol Cell Biol 2004; 24:8408 - 17; http://dx.doi.org/10.1128/MCB.24.19.8408-8417.2004; PMID: 15367662
- Piontek J, Winkler L, Wolburg H, Müller SL, Zuleger N, Piehl C, et al. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J 2008; 22:146 - 58; http://dx.doi.org/10.1096/fj.07-8319com; PMID: 17761522
- Overgaard CE, Mitchell LA, Koval M. Roles for claudins in alveolar epithelial barrier function. Ann N Y Acad Sci 2012; 1257:167 - 74; http://dx.doi.org/10.1111/j.1749-6632.2012.06545.x; PMID: 22671603
- Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 2005; 6:801 - 11; http://dx.doi.org/10.1038/nrm1736; PMID: 16314869
- Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 2005; 20:218 - 24; http://dx.doi.org/10.1152/physiol.00015.2005; PMID: 16024509
- Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys J 2006; 90:212 - 27; http://dx.doi.org/10.1529/biophysj.105.069666; PMID: 16352525
- Nusrat A, Parkos CA, Verkade P, Foley CS, Liang TW, Innis-Whitehouse W, et al. Tight junctions are membrane microdomains. J Cell Sci 2000; 113:1771 - 81; PMID: 10769208
- Lambert D, O’Neill CA, Padfield PJ. Methyl-beta-cyclodextrin increases permeability of Caco-2 cell monolayers by displacing specific claudins from cholesterol rich domains associated with tight junctions. Cell Physiol Biochem 2007; 20:495 - 506; http://dx.doi.org/10.1159/000107533; PMID: 17762176
- Van Itallie CM, Gambling TM, Carson JL, Anderson JM. Palmitoylation of claudins is required for efficient tight-junction localization. J Cell Sci 2005; 118:1427 - 36; http://dx.doi.org/10.1242/jcs.01735; PMID: 15769849
- Edidin M. The state of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct 2003; 32:257 - 83; http://dx.doi.org/10.1146/annurev.biophys.32.110601.142439; PMID: 12543707
- Kenworthy AK, Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J Cell Biol 1998; 142:69 - 84; http://dx.doi.org/10.1083/jcb.142.1.69; PMID: 9660864
- Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol 2011; 3:a004697; http://dx.doi.org/10.1101/cshperspect.a004697; PMID: 21628426
- Yamazaki Y, Okawa K, Yano T, Tsukita S, Tsukita S. Optimized proteomic analysis on gels of cell-cell adhering junctional membrane proteins. Biochemistry 2008; 47:5378 - 86; http://dx.doi.org/10.1021/bi8002567; PMID: 18416558
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 2007; 446:801 - 5; http://dx.doi.org/10.1038/nature05654; PMID: 17325668
- Kovalenko OV, Yang XH, Hemler ME. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol Cell Proteomics 2007; 6:1855 - 67; http://dx.doi.org/10.1074/mcp.M700183-MCP200; PMID: 17644758
- Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol. 1994;125(2):437-46. Epub 1994/04/01.
- Ladwein M, Pape UF, Schmidt DS, Schnölzer M, Fiedler S, Langbein L, et al. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp Cell Res 2005; 309:345 - 57; http://dx.doi.org/10.1016/j.yexcr.2005.06.013; PMID: 16054130
- Kuhn S, Koch M, Nübel T, Ladwein M, Antolovic D, Klingbeil P, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res 2007; 5:553 - 67; http://dx.doi.org/10.1158/1541-7786.MCR-06-0384; PMID: 17579117
- Wu CJ, Mannan P, Lu M, Udey MC. Epithelial Cell Adhesion Molecule (EpCAM) Regulates Claudin Dynamics and Tight Junctions. J Biol Chem 2013; http://dx.doi.org/10.1074/jbc.M113.457499; PMID: 23486470
- Nübel T, Preobraschenski J, Tuncay H, Weiss T, Kuhn S, Ladwein M, et al. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol Cancer Res 2009; 7:285 - 99; http://dx.doi.org/10.1158/1541-7786.MCR-08-0200; PMID: 19276185
- Thuma F, Zöller M. EpCAM-associated claudin-7 supports lymphatic spread and drug resistance in rat pancreatic cancer. Int J Cancer 2013; http://dx.doi.org/10.1002/ijc.28085; PMID: 23390083
- Lei Z, Maeda T, Tamura A, Nakamura T, Yamazaki Y, Shiratori H, et al. EpCAM contributes to formation of functional tight junction in the intestinal epithelium by recruiting claudin proteins. Dev Biol 2012; 371:136 - 45; http://dx.doi.org/10.1016/j.ydbio.2012.07.005; PMID: 22819673
- Daugherty BL, Ward C, Smith T, Ritzenthaler JD, Koval M. Regulation of heterotypic claudin compatibility. J Biol Chem 2007; 282:30005 - 13; http://dx.doi.org/10.1074/jbc.M703547200; PMID: 17699514
- Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol 1999; 147:891 - 903; http://dx.doi.org/10.1083/jcb.147.4.891; PMID: 10562289
- Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol 2003; 285:L1166 - 78; PMID: 12909588
- Blasig IE, Winkler L, Lassowski B, Mueller SL, Zuleger N, Krause E, et al. On the self-association potential of transmembrane tight junction proteins. Cell Mol Life Sci 2006; 63:505 - 14; http://dx.doi.org/10.1007/s00018-005-5472-x; PMID: 16456617
- Angelow S, Schneeberger EE, Yu AS. Claudin-8 expression in renal epithelial cells augments the paracellular barrier by replacing endogenous claudin-2. J Membr Biol 2007; 215:147 - 59; http://dx.doi.org/10.1007/s00232-007-9014-3; PMID: 17516019
- Sengoku A, Inai T, Shibata Y. Formation of aberrant TJ strands by overexpression of claudin-15 in MDCK II cells. Histochem Cell Biol 2008; 129:211 - 22; http://dx.doi.org/10.1007/s00418-007-0354-y; PMID: 17989991
- Nunes FD, Lopez LN, Lin HW, Davies C, Azevedo RB, Gow A, et al. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J Cell Sci 2006; 119:4819 - 27; http://dx.doi.org/10.1242/jcs.03233; PMID: 17130295
- Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 1999; 285:103 - 6; http://dx.doi.org/10.1126/science.285.5424.103; PMID: 10390358
- Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet 2006; 79:949 - 57; http://dx.doi.org/10.1086/508617; PMID: 17033971
- Hou J, Renigunta A, Gomes AS, Hou M, Paul DL, Waldegger S, et al. Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci U S A 2009; 106:15350 - 5; http://dx.doi.org/10.1073/pnas.0907724106; PMID: 19706394
- Angelow S, Yu AS. Claudins and paracellular transport: an update. Curr Opin Nephrol Hypertens 2007; 16:459 - 64; http://dx.doi.org/10.1097/MNH.0b013e32820ac97d; PMID: 17693762
- Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, et al. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest 2008; 118:619 - 28; PMID: 18188451
- Kausalya PJ, Amasheh S, Günzel D, Wurps H, Müller D, Fromm M, et al. Disease-associated mutations affect intracellular traffic and paracellular Mg2+ transport function of Claudin-16. J Clin Invest 2006; 116:878 - 91; http://dx.doi.org/10.1172/JCI26323; PMID: 16528408
- Koval M. Claudin Heterogeneity and Control of Lung Tight Junctions. Annu Rev Physiol 2013; 75:551 - 67; http://dx.doi.org/10.1146/annurev-physiol-030212-183809; PMID: 23072447
- Hou J, Renigunta A, Yang J, Waldegger S. Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc Natl Acad Sci U S A 2010; 107:18010 - 5; http://dx.doi.org/10.1073/pnas.1009399107; PMID: 20921420
- Surma MA, Klose C, Simons K. Lipid-dependent protein sorting at the trans-Golgi network. Biochim Biophys Acta 2012; 1821:1059 - 67; http://dx.doi.org/10.1016/j.bbalip.2011.12.008; PMID: 22230596
- Santos AX, Riezman H. Yeast as a model system for studying lipid homeostasis and function. FEBS Lett 2012; 586:2858 - 67; http://dx.doi.org/10.1016/j.febslet.2012.07.033; PMID: 22824640
- Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y, et al. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell 2010; 21:1200 - 13; http://dx.doi.org/10.1091/mbc.E09-08-0734; PMID: 20164257
- Steed E, Rodrigues NT, Balda MS, Matter K. Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol 2009; 10:95; http://dx.doi.org/10.1186/1471-2121-10-95; PMID: 20028514
- McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita S, Rogers RA, et al. Occludin is a functional component of the tight junction. J Cell Sci 1996; 109:2287 - 98; PMID: 8886979
- Singh U, Van Itallie CM, Mitic LL, Anderson JM, McClane BA. CaCo-2 cells treated with Clostridium perfringens enterotoxin form multiple large complex species, one of which contains the tight junction protein occludin. J Biol Chem 2000; 275:18407 - 17; http://dx.doi.org/10.1074/jbc.M001530200; PMID: 10749869
- Robertson SL, Smedley JG 3rd, Singh U, Chakrabarti G, Van Itallie CM, Anderson JM, et al. Compositional and stoichiometric analysis of Clostridium perfringens enterotoxin complexes in Caco-2 cells and claudin 4 fibroblast transfectants. Cell Microbiol 2007; 9:2734 - 55; http://dx.doi.org/10.1111/j.1462-5822.2007.00994.x; PMID: 17587331
- Van Itallie CM, Betts L, Smedley JG 3rd, McClane BA, Anderson JM. Structure of the claudin-binding domain of Clostridium perfringens enterotoxin. J Biol Chem 2008; 283:268 - 74; http://dx.doi.org/10.1074/jbc.M708066200; PMID: 17977833
- Winkler L, Gehring C, Wenzel A, Müller SL, Piehl C, Krause G, et al. Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J Biol Chem 2009; 284:18863 - 72; http://dx.doi.org/10.1074/jbc.M109.008623; PMID: 19429681
- Mitchell LA, Koval M. Specificity of interaction between clostridium perfringens enterotoxin and claudin-family tight junction proteins. Toxins (Basel) 2010; 2:1595 - 611; http://dx.doi.org/10.3390/toxins2071595; PMID: 22069652
- Mrsny RJ, Brown GT, Gerner-Smidt K, Buret AG, Meddings JB, Quan C, et al. A key claudin extracellular loop domain is critical for epithelial barrier integrity. Am J Pathol 2008; 172:905 - 15; http://dx.doi.org/10.2353/ajpath.2008.070698; PMID: 18349130
- Van Itallie CM, Mitic LL, Anderson JM. Claudin-2 forms homodimers and is a component of a high molecular weight protein complex. J Biol Chem 2011; 286:3442 - 50; http://dx.doi.org/10.1074/jbc.M110.195578; PMID: 21098027
- Cording J, Berg J, Kading N, Bellmann C, Tscheik C, Westphal JK, et al. Tight junctions: Claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J Cell Sci 2012; 126:554 - 64; http://dx.doi.org/10.1242/jcs.114306; PMID: 23203797
- Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 2005; 171:939 - 45; http://dx.doi.org/10.1083/jcb.200510043; PMID: 16365161
- Krug SM, Amasheh S, Richter JF, Milatz S, Günzel D, Westphal JK, et al. Tricellulin forms a barrier to macromolecules in tricellular tight junctions without affecting ion permeability. Mol Biol Cell 2009; 20:3713 - 24; http://dx.doi.org/10.1091/mbc.E09-01-0080; PMID: 19535456
- Shen L, Weber CR, Turner JR. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol 2008; 181:683 - 95; http://dx.doi.org/10.1083/jcb.200711165; PMID: 18474622
- Mitic LL, Unger VM, Anderson JM. Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci 2003; 12:218 - 27; http://dx.doi.org/10.1110/ps.0233903; PMID: 12538885
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 2003; 4:181 - 91; http://dx.doi.org/10.1038/nrm1052; PMID: 12612637
- Sitia R, Molteni SN. Stress, protein (mis)folding, and signaling: the redox connection. Sci STKE 2004; 2004:pe27; http://dx.doi.org/10.1126/stke.2392004pe27; PMID: 15226511
- Maza J, Mateescu M, Das Sarma J, Koval M. Differential oligomerization of endoplasmic reticulum-retained connexin43/connexin32 chimeras. Cell Commun Adhes 2003; 10:319 - 22; PMID: 14681035
- Das S, Smith TD, Sarma JD, Ritzenthaler JD, Maza J, Kaplan BE, et al. ERp29 restricts Connexin43 oligomerization in the endoplasmic reticulum. Mol Biol Cell 2009; 20:2593 - 604; http://dx.doi.org/10.1091/mbc.E08-07-0790; PMID: 19321666
- Laird DW. Life cycle of connexins in health and disease. Biochem J 2006; 394:527 - 43; http://dx.doi.org/10.1042/BJ20051922; PMID: 16492141
- Koval M. Pathways and control of connexin oligomerization. Trends Cell Biol 2006; 16:159 - 66; http://dx.doi.org/10.1016/j.tcb.2006.01.006; PMID: 16490353
- Das Sarma J, Wang F, Koval M. Targeted gap junction protein constructs reveal connexin-specific differences in oligomerization. J Biol Chem 2002; 277:20911 - 8; http://dx.doi.org/10.1074/jbc.M111498200; PMID: 11929864
- Gong XQ, Shao Q, Lounsbury CS, Bai D, Laird DW. Functional characterization of a GJA1 frameshift mutation causing oculodentodigital dysplasia and palmoplantar keratoderma. J Biol Chem 2006; 281:31801 - 11; http://dx.doi.org/10.1074/jbc.M605961200; PMID: 16891658
- Das Sarma J, Kaplan BE, Willemsen D, Koval M. Identification of rab20 as a potential regulator of connexin 43 trafficking. Cell Commun Adhes 2008; 15:65 - 74; http://dx.doi.org/10.1080/15419060802014305; PMID: 18649179
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 2009; 458:597 - 602; http://dx.doi.org/10.1038/nature07869; PMID: 19340074
- Piontek J, Fritzsche S, Cording J, Richter S, Hartwig J, Walter M, et al. Elucidating the pr. inciples of the molecular organization of heteropolymeric tight junction strands. Cell Mol Life Sci 2011;68:3903-18
- Smith TD, Mohankumar A, Minogue PJ, Beyer EC, Berthoud VM, Koval M. Cytoplasmic amino acids within the membrane interface region influence connexin oligomerization. J Membr Biol 2012; 245:221 - 30; http://dx.doi.org/10.1007/s00232-012-9443-5; PMID: 22722762
- Kaufmann R, Piontek J, Grüll F, Kirchgessner M, Rossa J, Wolburg H, et al. Visualization and quantitative analysis of reconstituted tight junctions using localization microscopy. PLoS One 2012; 7:e31128; http://dx.doi.org/10.1371/journal.pone.0031128; PMID: 22319608