Abstract
Three recent studies have investigated interspecies retention of binding sites of peroxisome proliferator-activated receptor γ (PPARγ), the master regulator of adipocyte differention, between mouse and human adipocytes. Here we discuss the major findings and demonstrate that retention of binding events is highly context-dependent.
Introduction
Adipocyte differentiation is controlled by activation of cascades of transcription factors that induce or repress the expression of each other in a sequential fashion.Citation1,Citation2 Recent studies of the genome-wide binding sites of adipogenic transcription factors have contributed with important novel insight into this transcriptional network. The master regulator of adipogenesis, the peroxisome proliferator-activated receptor γ (PPARγ), has received particular attention. First, studies showed that PPARγ binds in the vicinity of the majority of genes that are induced during adipogenesis, indicating that PPARγ is directly involved in the activation of most adipocyte genes. Second, induced genes have multiple binding sites of PPARγ indicating that a gene may be regulated by PPARγ through the concerted action of multiple binding sites. Third, although the proximal promoters of induced genes are enriched in PPARγ binding sites, most sites are far from the promoter, as approximately 50% of all sites are located in intragenic regions, and 40% are located at distant intergenic positions.Citation3-Citation5 Fourth, binding of another key adipogenic transcription factor, the CCAAT/enhancer binding protein α (C/EBPα), displays extensive overlap with PPARγ binding in mature adipocytes.Citation3,Citation6 Overlap between transcription factors has proven to be a common phenomenon.Citation7-Citation11 Recently, we demonstrated the existence of a large number of transcription factor hotspots at early time points of adipocyte differentiation to which multiple transcription factors bind in a cooperative manner,Citation9 and we consider it likely that similar hotspots exist and bind PPARγ at later stages of adipocyte differentiation.
The wealth of potential regulatory sites motivates strategies for finding the most important regulatory ones—i.e., are all of the sites equally important, or are some due to spurious binding? One such strategy is to investigate what sites are retained between species.Citation12-Citation18 While there is no mechanistic reason why species-specific sites are not functional, one can argue that highly conserved sites are subject to specific constraints and are therefore interesting. Such analyses will also give hints on the rigidity or plasticity of regulatory programs over evolution, and thus give insights on the regulatory strategies used by the cell in the process. Recently, the Rosen, Kaestner/Lazar and Mandrup/Sandelin laboratories all used ChIP-seq to study interspecies retention of PPARγ binding in mouse and human adipocytes.Citation6,Citation19,Citation20 Here, we summarize the findings made in these three studies and discuss how they provide novel insight into regulatory strategies for PPARγ in adipocytes.
Interspecies Comparison of PPARγ Binding
The three studies from the Rosen, Kaestner/Lazar and Mandrup/Sandelin laboratories all address interspecies retention of PPARγ between mouse and human adipocytes. All three studies used differentiated 3T3-L1 cells as mouse adipocyte model, and found between 7,000 and 13,000 PPARγ binding sites.Citation6,Citation19,Citation20 The difference in numbers of identified sites is most likely due to variations in experimental setup, such as cell culture conditions, peak detection methods and cutoffs. With respect to the human PPARγ binding sites, the Kaestner/Lazar group and we identified approximately 21,000Citation20 and 23,000Citation6 sites, respectively, using the human preadipocyte cell line Simpson Golabi Behmel Syndrome (SGBS). The Rosen group identified approximately 40,000 sites in primary in vitro differentiated human adipose stromal cells (hASC).Citation19 The numbers of PPARγ binding sites identified in both human adipocyte models are notably higher than in 3T3-L1 cells, which may reflect difference in antibody sensitivity toward human and mouse PPARγ and/or differences in chromatin accessibility between 3T3-L1 cells and the two human systems.
Retention of PPARγ Target Genes vs. Target Sites
All three studies focus on retention of mouse PPARγ binding sites in orthologous genomic regions in human. Thus, retention is not necessarily the same as evolutionary conservation at sequence level. Approximately 20–30% of PPARγ binding regions in the mouse genome are difficult to assign to a corresponding orthologous region in human based on sequence alignments. This may be due to large deletions or insertions; in addition, as noted in the work from the Rosen group, rodent-specific transposable elements account for a large fraction of these regions.Citation19
Focusing on regions that are clear orthologous pairs, only approximately 10–20% of mouse PPARγ sites are retained, i.e., the orthologous human region also contain a PPARγ binding site.Citation6,Citation19,Citation20 The slight differences between the studies in the percentage of retained sites most likely originate from differences in experimental setup and bioinformatic strategies. While the low retention of binding sites between mouse and human adipocytes may seem surprising, this low number is well in line with previous studies of other transcription factors in liverCitation13-Citation15 and embryonic stem cells (ESCs),Citation12 where 5–20% of binding sites are retained between mouse and human. Notably, whereas the position of PPARγ binding sites has diverged significantly between the two species, PPARγ target genes appear to be significantly more conserved than actual binding sites. The Kaestner/Lazar groups found that 50% of genes associated with nearby PPARγ binding in mouse adipocytes also had a nearby PPARγ binding site in human adipocytes.Citation20 Similarly, we found that the enrichment of PPARγ binding sites near genes upregulated during 3T3-L1 adipogenesis, was conserved near the orthologous human genes.Citation6 It is likely that this increased conservation of PPARγ target genes over binding site retention per se is due in part to binding site turnover, i.e., the compensation of a lost binding site with a gained nearby site. In line with this, we found that 25% of all mouse PPARγ binding sites are replaced by a species specific site within 10 kb of the loss.Citation6
Factors Influencing Target Site Retention
The low level of interspecies retention of binding sites indicates that functionally important binding sites cannot be identified solely based on retention. However, cross-species analyses of binding sites can be used to study what types of biological features influence retention. The three studies identified several such features in the mouse and human genomes that are correlated with retention of mouse binding sites in human and thus likely to reflect important aspects of PPARγ function (). Despite slightly different estimates of the fraction of PPARγ sites that are retained between species, there is agreement between the three studies regarding which factors influence retention.
Figure 1. Schematic views of features with a positive influence on retention of PPARγ binding. Retention is increased by proximity to induced genes,Citation6,Citation19 binding intensityCitation6,Citation20 H3K27ac in the orthologous human region,Citation19 co-occurrence with C/EBPα,Citation6 conservation of the predicted binding siteCitation6,Citation19,Citation20 and overall sequence conservation.Citation6,Citation20
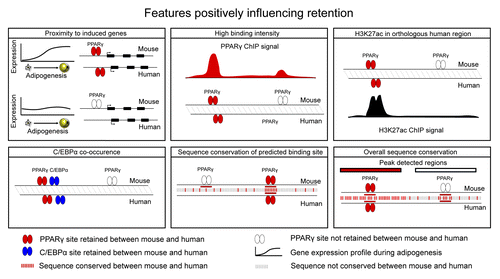
Sites in the vicinity of adipocyte genes display increased retention
By assigning PPARγ sites to the closest TSS within 100 kb, we found that retention is increased for binding sites in the vicinity of genes upregulated during 3T3-L1 adipogenesis.Citation6 Similarly, Rosen and colleagues approached the same question from a different angle and found that genes with nearby retained PPARγ sites are more likely to be upregulated during adipogenesis than genes in the vicinity of species specific PPARγ site,Citation19 although species specific sites are also likely to regulate nearby genes.Citation19,Citation20 Thus, selective pressure on transcription factor binding sites appears to be increased in the vicinity of putative target genes. Interestingly, turnover of PPARγ binding is also increased in the vicinity of genes upregulated during adipogenesis,Citation6 indicating that both conservation of sites and turnover contributes to the overall retention of the regulatory program between species. For instance, in the vicinity of genes highly upregulated during adipogenesis, > 60% of mouse PPARγ binding sites are either retained or undergo turnover.Citation6
Sequence conservation of larger regions has a high influence on retention
As expected, retention of PPARγ binding is dependent on sequence conservation of the PPARγ binding sites, albeit we and the Rosen group found that conservation of a PPARγ binding site sequence is not sufficient for retention of PPARγ binding.Citation6,Citation19 The Kaestner/Lazar group also reported a strong correlation between motif conservation and retention; however interestingly, regions with loss of PPARγ binding showed enrichment of the predicted PPARγ binding sites compared with random genomic regions.Citation20 Thus, loss of binding occurs even when a bona fide sequence element is present in both species, indicating that other features than the actual sequence that PPARγ binds to are important. This is consistent with the low information contained in binding motifs in general.Citation21 Interestingly, retention of PPARγ binding is increased in regions with high sequence identity between mouse and human within the larger binding regions (300–400 bp) identified by the peak detection software.Citation6 However surprisingly, conservation of predicted PPARγ binding sites within those regions are not higher than in other regions,Citation6 indicating that in this case it is the conservation of the broader regions and not the specific PPARγ-bound pattern that has an impact on the retention. This in turn suggests that in many of these regions, PPARγ binding is controlled by other sequence patterns with higher evolutionary constraints; some of which might be recognition motifs for other DNA-binding proteins. In line with our findings, Kaestner/Lazar and colleagues found that retained PPARγ sites have increased phastCon scores, i.e., have higher sequence conservation, compared with non-retained sites.Citation20
Overlap with C/EBPα binding increases retention
To investigate whether the overlap between PPARγ and C/EBPα, which was originally detected in 3T3-L1 adipocytes, was conserved in human adipocytes, we performed C/EBPα ChIP-seq in SGBS adipocytes.Citation6 The results showed this overlap is at least as extensive in human SGBS adipocytes as in 3T3-L1 cells.Citation6 Intriguingly, retention of both PPARγ and C/EBPα binding is increased at sites bound by both factors in mice, and shared binding sites are also primarily retained as shared binding sites.Citation6 Thus, the extensive overlap between PPARγ and C/EBPα binding appears to be of such functional significance, that shared binding sites experience increased selective pressure. Furthermore, analysis of the underlying sequence properties showed that the C/EBP binding motif hits are indicative of retention of PPARγ, but not vice versa, suggesting that C/EBPα facilitates PPARγ binding to DNA.Citation6 Similar to our findings, Hemberg and Kreiman recently showed that retention of transcription factor binding between mouse and human is increased in regions bound by multiple transcription factors in liver compared with regions bound by only one of the investigated transcription factors.Citation22
Histone marks influence retention
In addition to profiling transcription factors, Rosen and colleagues profiled various histone modifications at different time points throughout mouse and human adipogenesis.Citation19 Similar to the retention of transcription factor binding, they found that between 15–30% of regions enriched for a histone mark in mouse 3T3-L1 cells contains the same mark in the orthologous region in human adipocytes. Moreover, species-specific active histone marks correlate with species specific gene expression of nearby genes.Citation19 In both species, PPARγ binds predominantly (85–95%) to regions containing one or more of the histone modifications H3K4me, H3K4me2 or H3K27ac, which are typically associated with open chromatin.Citation19 Consequently, retention of mouse PPARγ binding correlates strongly with the presence of active chromatin marks in the orthologous human region.Citation19 Notably, 77% of all mouse PPARγ binding sites are located in regions that already in the preadipocyte state contain H3K4me, H3K4me2 or H3K27ac, suggesting that the chromatin state in preadipocytes strongly influences PPARγ binding in mature adipocytes.Citation19
Interestingly, the Rosen group found that genes associated with PPARγ binding sites are more likely to be upregulated during adipogenesis if the H3K27ac signal at the site increases more than 5 fold over the course of adipogenesis.Citation19 This indicates that whereas PPARγ is likely to bind to regions that are open already in the preadipocyte state, PPARγ sites that gain acetylation during adipogenesis are more likely to be involved in gene regulation. Given the indicated importance of these “dynamic” PPARγ binding sites, we hypothesized that they are more likely to be retained between species. Using data from the Rosen studyCitation19 and the 5 fold change in H3K27ac during adipogenesis as cutoff, we divided mouse PPARγ sites into “dynamic” and “nondynamic“ sites and investigated the mouse-human retention of these two groups as described in the Mandrup/Sandelin studyCitation6 (). Surprisingly, the “dynamic” PPARγ sites are less likely to be retained in humans, despite the stronger correlation of these sites with gene regulation during mouse adipogenesis.Citation19
Figure 2. Correlation between H3K27ac and retention of PPARγ binding sites. The analyses were performed using publically available PPARγ and H3K27ac data from the Rosen groupCitation19 (GEO accession GSE20752) and DHS data from the Mandrup groupCitation9 (GEO accession GSE27826). For each PPARγ peak (GSM535769), mean H3K27ac tag/10e6 and DHS tag/10e6 counts were calculated for preadipocytes and adipocytes. Retention of PPARγ sites were calculated as described in the Mandrup/Sandelin study.Citation6 (A) Bar diagram representing the fraction of retained sites for “dynamic” and “nondynamic” PPARγ binding sites. PPARγ peaks with > 5 fold more H3K27ac tags in adipocytes than in preadipocytes were considered dynamic. *p = 7.3*10^-17(Fischer Exact test). (B) Heatmap showing the fraction of retained sites for mouse PPARγ binding sites divided into 16 groups based on their acetylation levels in pre-adipocytes and adipocytes, respectively. The groups are 0–10, 10–20, 20–40 and > 40 tags pr 10e.Citation6 The color scale goes from red (low) retention to white (high) retention. (C+D) Box-whisker plots representing H3K27ac tag counts (C) and DHS tag counts (D) for “dynamic” and “nondynamic” PPARγ binding sites in preadipocytes. #p < 2.2*10^-16 (Wilcoxon test).
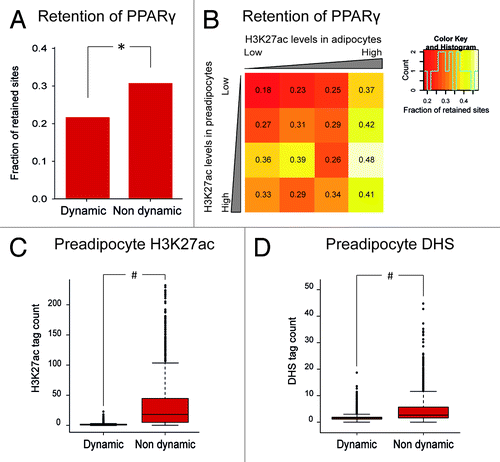
To further investigate the correlation between H3K27ac and retention of PPARγ binding, we divided mouse PPARγ sites into groups based on their H3K27ac levels in both preadipocytes and adipocytes and calculated retention of PPARγ sites in these groups (). Interestingly, PPARγ binding sites with low acetylation in both preadipocytes and adipocytes have low retention, whereas PPARγ sites are more likely to be retained, if they have high acetylation levels in either preadipocytes, adipocytes or both. Thus, the total H3K27ac levels in the PPARγ binding regions seem to be more important for retention than the development of acetylation during adipogenesis. Although the causal relationship between H3K27ac levels and retention of PPARγ remains unclear, it is most likely related to chromatin accessibility and enhancer activity, two features associated with H3K27acetylated regions. Thus, regions with high acetylation levels are likely to be highly accessible and functionally important regulatory regions, which experiences high selective pressure. Overall, “non-dynamic” PPARγ sites have more H3K27ac in preadipocytes than “dynamic” sites () and have a more open chromatin structure in preadipocytes as assessed by DNase I hypersensitivity (DHS) assayCitation9 (). This suggests an important role for these regions in regulation of preadipocyte gene expression, which may explain the increased retention of “non-dynamic” PPARγ sites compared with “dynamic” sites.
Discussion
Evolutionary pressure on PPARγ binding per se or context dependent retention?
The comparative genome-wide analyses of PPARγ binding in mouse and human adipocytes showed that retention of binding sites is low, but clearly dependent on context, as a number of features, which positively affects retention have been identified (). Vicinity to highly expressed genes (probable target genes), co-binding with C/EBPα, and binding strength, are features of the mouse PPARγ sites which influences whether a binding site is likely to be retained in human or not, whereas features in the orthologous human regions that influence retention includes actual sequence conservation and chromatin context. In general, the latter are more strongly correlated with retention, but a clear ranking of these features is difficult due to the dependency of cutoffs in these analysis. Except for a small increase in retention of PPARγ sites which are both shared by C/EBPα and located in the vicinity of induced genes,Citation6 we have not found indications of strong additivity between the different features.
While correlation does not imply causation, it gives ample material for new hypotheses. It is clear that the context of the binding sites can be used to classify sites in different categories, which resist evolutionary changes with different rates. It is highly likely that evolutionary pressure on PPARγ binding per se accounts for increased retention close to genes induced during adipogenesis, because these PPARγ sites are involved in regulation of the genes.
Similarly, the increased retention at PPARγ sites with co-binding of C/EBPα may be caused by the higher evolutionary pressure on these sites due to the importance of crosstalk between trancription factors. On the other hand, the increased retention could also be a context effect, since the overall information content in a cis-regulatory module containing multiple different binding sites is harder to recreate (e.g., turnover) and therefore should stay more static over evolution.
It is interesting—and confusing—to note that the dynamics of H3K27ac at mouse PPARγ sites seems to be a reasonable predictor of whether the sites are important for regulation during adipogeneis, but not for the actual retention of the sites, whereas retention correlates with the intensity of the H3K27ac signal regardless of dynamics. An example is the increased retention of PPARγ sites with high acetylation levels in preadipocytes and low levels in adipocytes compared with sites with low acetylation levels at both stages. This indicates that high acetylation in the preadipocyte increases retention of PPARγ binding in the mature adipocyte. The reason for this is unclear, but we speculate that sites with high acetylation levels in mouse preadipocytes represent regions that experience high evolutionary pressure, because they play an active role in undifferentiated cells, independent of later PPARγ binding to these regions. Thus, the context of the binding site may significantly influence retention of PPARγ binding independent of the evolutionary pressure on PPARγ per se, indicating that cross-species ChIP-seq alone is not an optimal strategy for identification of the most functionally important PPARγ binding sites. However, by combining cross species ChIP-seq with H3K27ac profiling during adipogenesis, Rosen and colleagues were able to identify a group of retained and “dynamic” PPARγ sites with stronger correlation to induction of gene expression during adipogenesis than any of these two criteria alone.Citation19
Importance of non-retained sites?
Similar to what has been found in cross-species analyses of other transcription factors, the majority of mouse PPARγ binding sites are not retained in human adipocytes. It is important to stress that this does not imply that the majority of PPARγ binding sites are unimportant, although it is easy to envision a subclass of PPARγ sites which are binding to accessible DNA and do not have a direct influence on a target gene. Most target genes seem to have an array of sites where many are species-specific on site level but not on gene-target level. Related to this, genes associated with species-specific PPARγ binding sites are more likely to be upregulated during adipogenesis than genes with no nearby PPARγ binding sites.Citation19 In addition, genes that are associated with PPARγ sites in mouse only, are more likely to be affected by siRNA-mediated knockdown of PPARγ than genes with no associated PPARγ sites.Citation20 The reason for having many sites close to target genes is unknown, and at present we know close to nothing about the relative importance of these sites for any single gene. It is conceivable, that sites play either a redundant role where one site can fully take over from another, or that the expression of a gene is a sum of contributions from multiple sites in collaboration. Both of these scenarios would allow for a large evolutionary plasticity and the possibility to fine-tune expression by moving, removing or adding additional sites gradually without lethal effects.
Future challenges
It is clear that cross-species retention is not in itself a sensitive tool for identifying functional sites, and experimental approaches are needed to link binding events to function, i.e., determine the relative importance of the binding sites of a particular factor with respect to regulation of potential target genes. Fine-grained time series combining ChIP-seq and expression profiling is one approach, but links remain correlative. Other avenues that offer complementary angles include chromatin conformation capture experiments that can link binding events to actual core promoter regions.Citation23,Citation24 Yet these do not prove function. The determination of causalities requires removal of sites followed by transcriptional analysis. This is a very difficult approach, not just because of the large number of binding sites and the tedious and expensive targeting strategies, but also because transcription factors may bind in a cooperative way to hotspots,Citation9 making it difficult to determine the importance of one factor without interfering with the binding of multiple others. Lastly, it will be important to determine how different co-binding factors impact on the ability of PPARγ to engage in gene activation and/or gene repression and how these factors may influence the ability of PPARγ to respond to agonists.
| Abbreviations: | ||
| C/EBP | = | CAAT/enhancer binding protein |
| ChIP | = | chromatin immunoprecipitation |
| DHS | = | DNase I hypersensitivity |
| ESC | = | embryonic stem cells |
| hASC | = | human adipose stromal cells |
| PPAR | = | peroxisome proliferator activated receptor |
| SGBS | = | Simpson Golabi Sehmel syndrom |
| TSS | = | Transcription start site |
Acknowledgments
The authors thank members of the Mandrup and Sandelin laboratories for valuable discussions and input to the manuscript. This work was supported by grants to S.M. from the Novo Nordisk Foundation and the NIH NIDDK/ODS (5R01-DK063070) and by grants from European Research Council (FP7/2007–2013/ERC grant agreement 204135), the Novo Nordisk Foundation and the Lundbeck Foundation to A.S.
References
- Siersbæk R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab 2011; PMID: 22079269
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 2011; 12:722 - 34; http://dx.doi.org/10.1038/nrm3198; PMID: 21952300
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 2008; 22:2941 - 52; http://dx.doi.org/10.1101/gad.1709008; PMID: 18981473
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev 2008; 22:2953 - 67; http://dx.doi.org/10.1101/gad.501108; PMID: 18981474
- Siersbaek R, Nielsen R, Mandrup S. PPARgamma in adipocyte differentiation and metabolism--novel insights from genome-wide studies. FEBS Lett 2010; 584:3242 - 9; http://dx.doi.org/10.1016/j.febslet.2010.06.010; PMID: 20542036
- Schmidt SF, Jørgensen M, Chen Y, Nielsen R, Sandelin A, Mandrup S. Cross species comparison of C/EBPα and PPARγ profiles in mouse and human adipocytes reveals interdependent retention of binding sites. BMC Genomics 2011; 12:152; http://dx.doi.org/10.1186/1471-2164-12-152; PMID: 21410980
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A 2011; 108:5632 - 7; http://dx.doi.org/10.1073/pnas.1016959108; PMID: 21415370
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133:1106 - 17; http://dx.doi.org/10.1016/j.cell.2008.04.043; PMID: 18555785
- Siersbæk R, Nielsen R, John S, Sung MH, Baek S, Loft A, et al. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J 2011; 30:1459 - 72; http://dx.doi.org/10.1038/emboj.2011.65; PMID: 21427703
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 2010; 24:1035 - 44; http://dx.doi.org/10.1101/gad.1907110; PMID: 20478996
- Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesbøll C, et al. Genome-wide profiling of LXR, RXR and PPARα in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 2011; http://dx.doi.org/10.1128/MCB.06175-11; PMID: 22158963
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 2010; 42:631 - 4; http://dx.doi.org/10.1038/ng.600; PMID: 20526341
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 2007; 39:730 - 2; http://dx.doi.org/10.1038/ng2047; PMID: 17529977
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 2010; 328:1036 - 40; http://dx.doi.org/10.1126/science.1186176; PMID: 20378774
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, et al. Species-specific transcription in mice carrying human chromosome 21. Science 2008; 322:434 - 8; http://dx.doi.org/10.1126/science.1160930; PMID: 18787134
- Wilson MD, Odom DT. Evolution of transcriptional control in mammals. Curr Opin Genet Dev 2009; 19:579 - 85; http://dx.doi.org/10.1016/j.gde.2009.10.003; PMID: 19913406
- Johnson R, Samuel J, Ng CK, Jauch R, Stanton LW, Wood IC. Evolution of the vertebrate gene regulatory network controlled by the transcriptional repressor REST. Mol Biol Evol 2009; 26:1491 - 507; http://dx.doi.org/10.1093/molbev/msp058; PMID: 19318521
- Dowell RD. Transcription factor binding variation in the evolution of gene regulation. Trends Genet 2010; 26:468 - 75; http://dx.doi.org/10.1016/j.tig.2010.08.005; PMID: 20864205
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010; 143:156 - 69; http://dx.doi.org/10.1016/j.cell.2010.09.006; PMID: 20887899
- Soccio RE, Tuteja G, Everett LJ, Li Z, Lazar MA, Kaestner KH. Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol Endocrinol 2011; 25:694 - 706; http://dx.doi.org/10.1210/me.2010-0454; PMID: 21292830
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 2004; 5:276 - 87; http://dx.doi.org/10.1038/nrg1315; PMID: 15131651
- Hemberg M, Kreiman G. Conservation of transcription factor binding events predicts gene expression across species. Nucleic Acids Res 2011; 39:7092 - 102; http://dx.doi.org/10.1093/nar/gkr404; PMID: 21622661
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 2009; 462:58 - 64; http://dx.doi.org/10.1038/nature08497; PMID: 19890323
- Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 2007; 4:895 - 901; http://dx.doi.org/10.1038/nmeth1114; PMID: 17971780