Figures & data
Table 1 Patient demographics, bacterial strains and features of plasmids carrying blaNDM-1
Figure 1 Restriction analysis of IncX3 plasmids carrying blaNDM-1. Plasmids were digested with (A) EcoRI and (B) PstI and separated by electrophoresis in 1% agarose. M, GeneRulerTM DNA ladder. The labels above each lane show the strain number, bacterial species origin (EA, E. aerogenes; KP, K. pneumoniae; EC, E. coli; EO, E. cloacae; CF, C. freundii) and the geographic source of importation (HN, Hunan; HF, Haifeng; DG, Dongguan; GZ, Guangzhou).
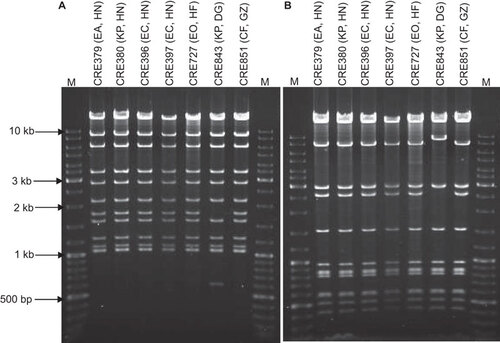
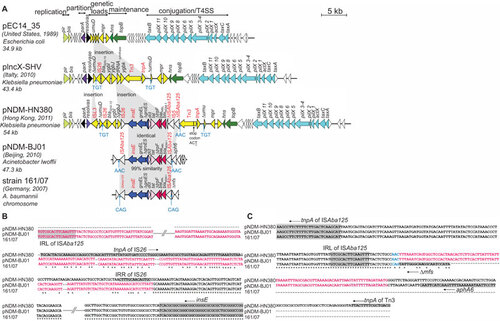
Figure 2 Comparative analysis of (A) linear plasmid maps for three IncX3 plasmids, pEC14_35, pIncX-SHV, pNDM-HN380 and two blaNDM-1-carrying transposon sequences in pNDM-BJ01 and A. baumannii strain 161/07; (B) sequences downstream of insE and (C) sequences upstream of the ISAba125 in the 5′ end of blaNDM-1. The function blocks of the plasmids are indicated above the linear maps. The lengths of the ORFs are drawn in proportion to the size of the ORFs. Homologous ORFs in the plasmid maps are represented in the same colour. Direct repeats and mobile elements are labelled in blue and red, respectively. (B, C) Consensus regions in the aligned sequences of pNDM-HN380, pNDM-BJ01 and 161/07 are marked with asterisk. The sequences identical in pNDM-HN380 and pNDM-BJ01 are coloured green. The ORFs are indicated by grey shading and the arrow next to the label indicates the ORF orientation. The accession numbers were: pEC14_35 (JN935899), pIncX-SHV (JN247852), pNDM-HN380 (JX104760), pNDM-BJ01 (JQ001791) and Acinetobacter baumannii strain 161/07 (HQ857107).