Figures & data
Figure 1 Schematic representation of RNAP structural elements including the RIF resistance determining region (RRDR). The cartoon showing the RNAP holoenzyme is adapted from Borukhuv and Nudler.Citation17 Structural annotations have been simplified, and the promoter sequence has been excluded. The rpoB-encoded β subunit is highlighted in green. A yellow star represents the RNAP active site and a red circle denotes the RIF molecule which approaches within 12 Å of the active site,Citation18 inhibiting transcription. Double-stranded DNA is represented by pink lines and, once unwound, only template DNA is shown, with the growing RNA chain colored in blue. The inset shows a simplified depiction of the RIF binding pocket.Citation18 Amino acids that form hydrogen bonds with RIF are highlighted in blue and those that form van der Waals interactions are colored yellow; amino-acid numbering corresponds to that used for E. coli. Mutations identified in 11 of the 12 residues that surround the RIF binding pocket have been associated with RIF resistance, albeit at different frequenciesCitation18 (the sole amino acid, E565, which has not been associated with RIFR mutations is colored in grey). A schematic representation of the rpoB gene which encodes the β subunit of RNA polymerase is shown below the RNAP cartoon (adapted from Campbell et al.Citation18). Amino-acid numbering is shown as dashed demarcations. The RRDR is highlighted in blue and the amino-acid sequence of the RRDR is magnified below. The alignment contains the amino-acid sequences of E. coli, T. aquaticus and M. tuberculosis. Amino acids that interact directly with RIF are indicated by circles and the colors correspond to the inset diagram. Circles highlighted in red indicate residues that are most frequently observed in RIFR isolates.Citation18
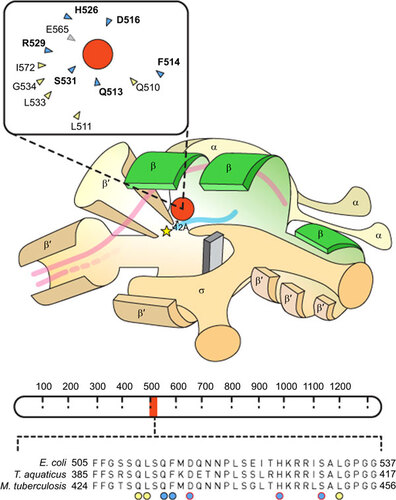
Table 1 Impact of RIFR-associated rpoB mutations on bacterial physiology
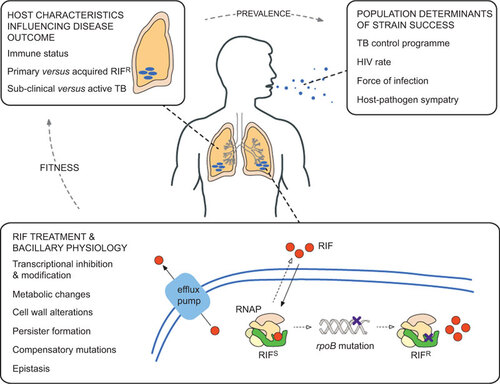
Figure 2 Factors influencing the success of RIFR Mtb strains. Although the focus of this review is on RIF resistance in Mtb, many of the themes are relevant to other drugs and other infectious organisms. RIF treatment and bacillary physiology: within a single bacterial cell, many factors contribute to the development and maintenance of drug resistance. The drug (in this case, RIF) enters the cell by passive diffusion and, once in the cytoplasm, must translocate and bind to its target (here, RNAP). Some organisms encode enzymes that inactivate RIF,Citation147 while recent work suggests that RIF is actively extruded by efflux pumps in Mtb.Citation112,Citation148 The concentration of RIF that is available to bind to RNAP (that is, the effective intracellular concentration) is a major determinant of whether resistance mutations develop or not, and is heavily influenced by the mechanisms described above;Citation149 therefore, the ability to measure this parameter accuratelyCitation150 will be critical to future efforts to understand the development of drug resistance. As described in the main text, mutations in rpoB might alter the physiology of the RIFR bacterium. Host characteristics influencing disease outcome: any physiological alteration has the potential to influence the interaction of the bacillus with its obligate human host. Similarly, multiple host factors such as age, nutritional status and copathologies will determine infection outcomes, including transmission to other susceptible individuals. Population determinants of strain success: there is an additional layer of complexity when considering the spread of organisms within and between host populations. Factors such as HIV prevalence, force of infection and socioeconomic status will influence the ability of the organism to transmit between hosts.Citation151 While these elements are grouped separately in the figure, it is likely that multiple factors will overlap to influence the success of different Mtb strains.