Figures & data
Figure 1 Phylogenetic analysis of the NS3 protease domain of diverse members of the hepaciviruses and pegiviruses (), with susceptible host species incorporated. Radial tree topology highlighting distinct viral clades (boxed) within the hepacivirus (top) and pegivirus (bottom) genera. Viral clades are color-coded according to susceptible host species (primates: red, horses: green, bats: blue and rodents: orange). Phylogeny was generated using 54 nucelotide sequences encoding a 534-bp fragment at the N-terminus of the NS3 gene, corresponding to the virally encoded protease domain (genome coordinates 3420–3954 HCV genotype 1a strain; accession number NC_004102). Sequences were aligned according to overlying amino acids using the Clustal W algorithm implemented in MEGA5, and gap-stripped prior to tree construction. Tree was generated using the maximum likelihood method implemented in MEGA5 under the GTR+I+Γ model of nucleotide substitution. Values assigned to deep internal nodes within the phylogeny represent bootstrap supports for groupings. Values presented are percentages, derived from 1000 replications, with only significant values shown (>70%). The pegivirus grouping and the deep divergences within it are generally well supported, indicating the monophyly of the genus. The grouping of HCV, NPHV/CHV and BHV-C and D within the hepacivirus genus is also well supported. However, the deep divergences apparent between the remaining members of the genus are characterised by extremely long connecting branch-lengths and non-significant bootstrap values. This is indicative of ancient divergence between members, in addition to sparseness of taxon sampling: it is likely that many hepaciviruses in this portion of the tree remain either undiscovered, or have become extinct along with their hosts. Branch lengths are in accordance with the scale bar and are proportional to nucleotide substitutions per site. BPgV* clade containing the virus formerly designated GBV-D. Bat hepacivirus, BHV; bat pegivirus, BPgV; rodent hepacivirus, RHV; rodent pegivirus, RPgV; guereza hepacivirus, GHV; Theiler's disease-associated virus, TDAV; equine pegivirus, EPgV.
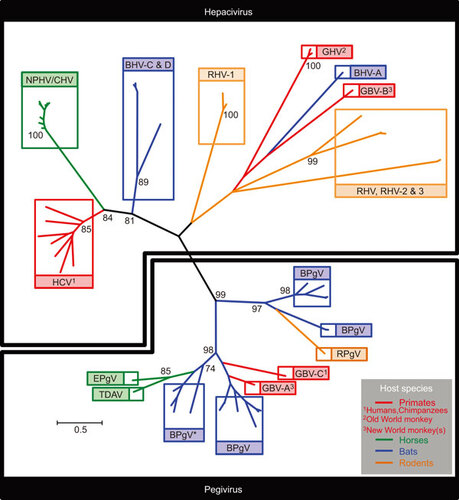
Table 2 The newly discovered hepaciviruses, host species, sequence accession number and isolate designation
Table 1 Comparison of features from novel hepaciviruses
Figure 2 Reference cartoons depicting polyprotein organisation and cleavage site coordinates for distinct hepaciviruses. Cartoons of recently described hepaciviral polyproteins relative to an HCV reference (bottom) are shown, depicting locations of homologous protease cleavage sites, with names of represented hepaciviruses located to the left of each polyprotein schematic. Coordinates located above each cartoon represent the first residue of each encoded protein. The 10 cleaved proteins are detailed below the schematic cartoon of the HCV genotype 1a reference polyprotein. Black and grey triangles located below the HCV cartoon indicate host and viral protease cleavage sites, respectively. Peptide cleavage sites have been experimentally validated for HCV, while recently identified hepaciviral homologs have predicted cleavage sites which require further experimental validation. Color coding according to susceptible host species is identical to Figure . Structural proteins (core, E1 and E2) associated with infectious virions are shaded dark, non-structural proteins essential for assembly (p7 and NS2) are shaded intermediate and non-structural proteins associated with the replication module (NS3, NS4A, NS4B, NS5A and NS5B) are shaded light. GenBank accession numbers for representative strains are HCV-1a: NC_004102, NPHV: JQ434008, CHV: JF744991, RHV: KC815310, RHV-1: KC411777, RHV-2: KC411784, RHV-3: KC411807, BHV-A: KC796077, BHV-C: KC796090 and BHV-D: KC7960074.
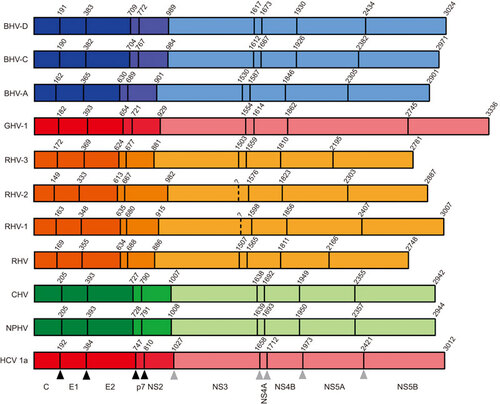
Table 3 The newly discovered pegiviruses, host species, sequence accession number and isolate designation
