Figures & data
Figure 1 (A) Sequence alignment of IFNLs. An amino-acid sequence alignment of human IFNL1–4 is shown. Red letters indicate significant differences. Green letters indicate common cysteines. Helices and exons are indicated with boxes. (B) Sequence alignment overlay for different IFNLs. Amino acids of significance are highlighted.
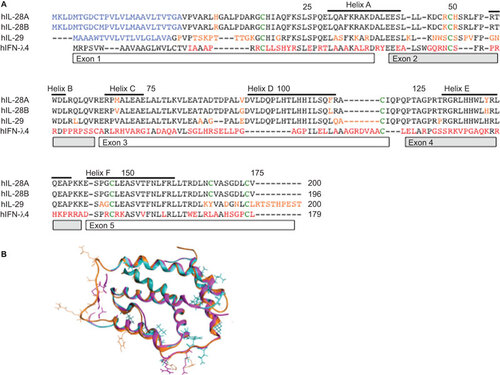
Figure 2 Expression dynamics of IFNs and ISGs during viral infection or inflammatory stimulation. Red line represents IFNL3, blue line IFN-α and black line IFNL4 expression dynamics. The lower panel illustrates the amount of ISG expression.
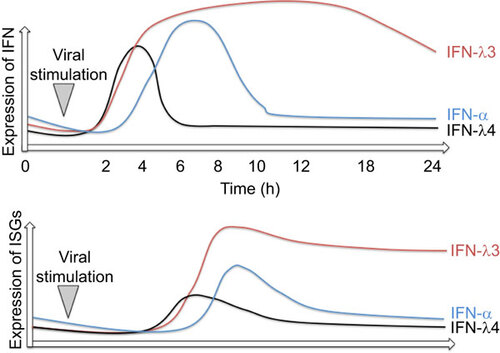
Table 1 Selection of important SNPs in the IFNL signaling cascade
Figure 3 Hypothesis of IFNL3 single nucleotide polymorphism and impact on virus replication. The left panel shows a major allele genotype with a presumed fully functional IFNL3 expression during virus replication. The right shows a minor allele genotype with reduced IFNL3 expression, but increased IFNL4 expression. The genotype results in a significantly different ISG background expression which may be an advantage or disadvantage for different viruses.
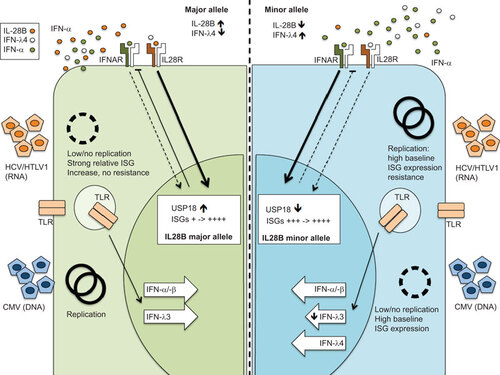
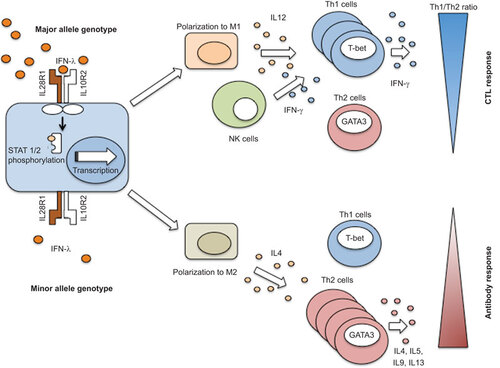
Figure 4 Impact of IFNL on Th1/Th2 cytokine profiling. The IFNL3 major allele genotype shows a predominant polarization to a macrophage M1 phenotype, whereas the minor allele genotype may show a relative increase of M2 phenotype. These macrophage phenotypes are associated with the downstream regulation and polarization of Th1 and Th2 cells. NK cells, natural killer cells.