Figures & data
Figure 1 Annual isolation of all avian influenza subtypes in poultry at different live-bird markets and farms in Bangladesh from August 2010 to December 2013. The total number of samples collected each year were 1198 (2010), 4897 (2011), 3952 (2012) and 2593 (2013).
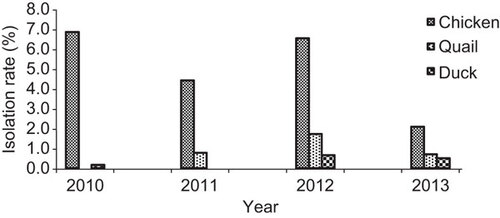
Figure 2 Monthly isolation of all H9N2 and H5N1 subtypes in poultry at different live-bird markets and farms in Bangladesh from August 2010 to December 2013.
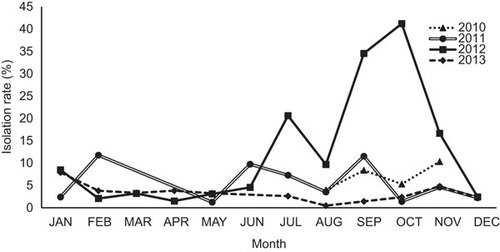
Figure 3 Nucleotide identity matrix of individual genes from Bangladeshi H9N2 viruses. The sequence identity range is indicated in different colors and above each gene segment. The arrow shows the earliest to the latest isolation date from 2010 to 2013. Colors indicate high (green, 99%–100%), median (yellow, 96%–98%) and low relative similarities (red, 94%–95%) for each indicated gene after the sequence identity was normalized in the matrix to have a mean of 0 and variance of 1.
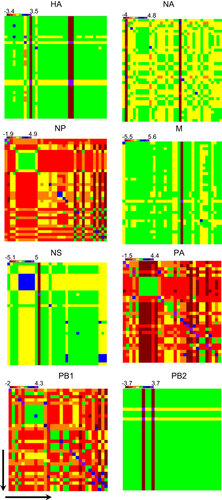
Figure 4 Unrooted maximum-likelihood phylogenetic trees for the (A) HA, (B) NA, (C) M, (D) NP, (E) NS, (F) PA, (G) PB1 and (H) PB2 genes of all analyzed H9N2 viruses. The trees were generated by minimum evolution analysis with maximum likelihood, using the Tamura-Nei model in MEGA (version 5.1). Full-length sequences with complete open reading frames were used for the phylogenetic analyses. Bootstrap values from 1000 replicates are indicated below the branches, and bootstrap values greater than 60 are shown. The scale bar represents the distance units between sequence pairs. Representative prototypes viruses from different H9N2 lineages are indicated in red, the human H9N2 isolate from Bangladesh is indicated in fuchsia and the H7N3 isolate from Pakistan is indicated in blue. The possible H5N1 recombinant isolates in the PB1 gene are indicated in green.
Figure 5 (A) Map showing the hypothesized and proposed routes taken by avian influenza H9N2 virus from Hong Kong to Bangladesh, based on H9N2 isolated from specific locations and years. Numbers 1–6 represent the sequence of events that took place after H9N2 was identified in Hong Kong in 1997.
H9N2 spread from Hong Kong to the United Arab Emirates (UAE; Middle East) and Pakistan and from Pakistan to Japan during and after 1997 via pet trade.
After establishing a separate lineage, H9N2 spread from the UAE to Pakistan during early 2000.
The Iranian H9N2 lineage moved to Pakistan via the cross-border poultry trade.
During 2004, H7N3/H9N2 reassortment occurred in Pakistan, followed by H9N2/H7N3 reassortment involving H9N2 lineages from the UAE and Iran.
H9N2/H7N3 reassortants spread to terrestrial poultry in India via the cross-border trade.
H9N2/H7N3 reassortants spread from India to Bangladesh, became firmly established and evolved into a distinct sublineage. (B) Evolution of Bangladeshi H9N2 through inter- and intrasubtypic reassortment between different H9N2 lineages and HPAI H7N3 and HP H5N1. H9N2 and H7N3 viruses are represented by black and red ovals, with each containing horizontal bars that represent the eight viral gene segments (starting from the top: PB2, PB1, PA, HA, NP, NA, M, NS). To illustrate the gene ancestry through reassortment events, the gene segments in descendent H9N2 viruses are colored according to their corresponding source viruses. Mideast B and C lineages of H9N2 were defined by lineage assignment by extended learning (LABEL).Citation17 In 1997, the prototypical H9N2 belonging to the G1 clade was identified in Hong Kong, and this lineage continued to circulate in poultry species until 2000. During 2000–2003, in the UAE and Iran, H9N2 formed distinct lineages with the G1 background that were identified by phylogenetic analyses. In 2004, reassortment occurred between H9N2/G1 and H7N3, thereby generating several true and hypothetical reassortants (broken oval). In 2004, H9N2/G1 also reassorted with H5N1 (green) to generate an H9N2 reassortant with the NS gene derived from H5N1. From 2009, many inter- and intrasubtypic reassortants between the H9N2 lineages from Dubai and Iran with H7N3 from Pakistan emerged to form several H9N2 influenza virus genotypes that moved through India to Bangladesh.

Table 1 H9N2 influenza viruses isolated from live-bird markets in Bangladesh from August 2010 to December 2013
Table 2 Nucleotide sequence homology of Bangladeshi H9N2 isolates and representative H9N2 viruses
