Figures & data
Figure 1 Route of the family traveling from an affected area to South China in late February 2016. On their way from Venezuela to Guangzhou, they had a layover in New York, where they stayed for four days.
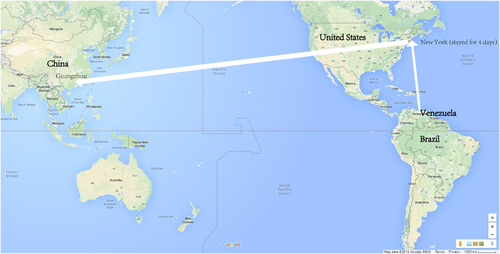
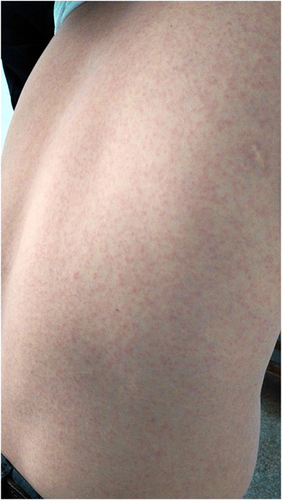
Figure 3 Inferred time axis ranging from probable biting by mosquitoes to release of ZIKV-infected family members. +, ++, +++ represent varying degree of rashes on the neck and back of patients. ▾ Time point when peripheral blood was positive for ZIKV using real-time PCR. ♦ Time point when urine or saliva was positive for ZIKV using real-time PCR. March 9 was the date of release from isolation.

Table 1 The clinical features and laboratory tests from the patients with ZIKV infection
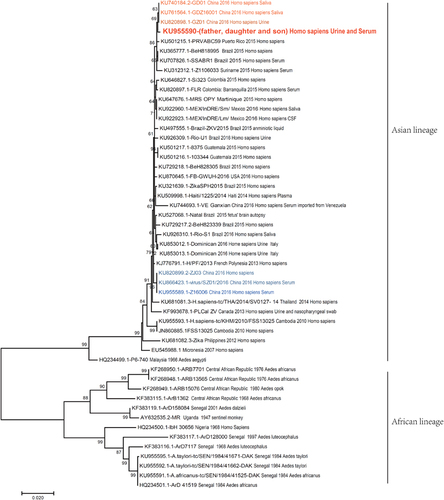
Figure 4 Phylogenetic tree based on E gene sequences of Zika virus isolates. E gene sequences from the father, daughter and son (Genbank: KU955590) in our study were used for alignment with other reference sequences. All isolates are indicated with related information (Genbank number, country, year and host, some with sample type). Phylogenetic trees were drawn using the maximum likelihood method by the Tamura-Nei model with gamma-distributed evolutionary rates in MEGA 7.0. An initial tree was made automatically with the nearest-neighbor-interchange (NNI) method. Gaps/missing data treatment was set as complete deletion. Bootstrap analyses with 1000 replications were utilized to determine confidence values for groupings within the phylogenetic trees. Other parameters were set to default style.