Figures & data
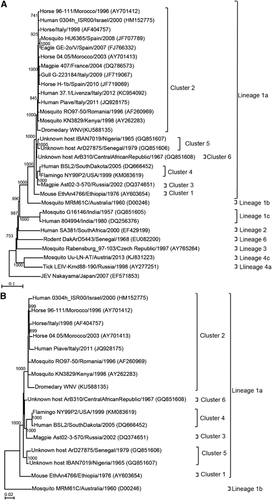
Figure 1 (A) Cytopathic effects of WNV from a dromedary on Vero cells, showing cell rounding, degeneration and detachment, magnification: × 40. (B) Negative contrast electron microscopic examination of an infected Vero cell culture supernatant showing enveloped icosahedral viral particles, bar=100 nm.
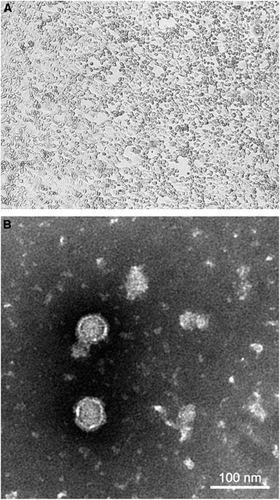
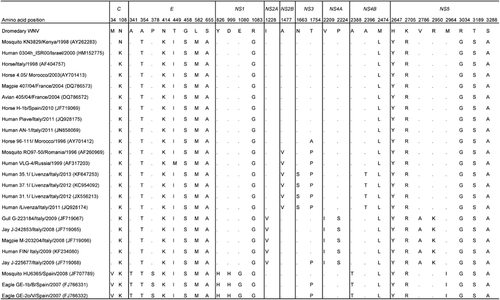
Figure 2 Phylogenetic tree showing the relationship of dromedary WNV to other WNV strains based on a complete polyprotein sequence by the maximum-likelihood method in MEGA5. (A) WNV strains of all lineages and clusters are indicated except for those of lineages 4b, 5 and 7 because no complete polyprotein sequence is available from these lineages. A total of 10 345 nucleotide positions were included in the analysis. Japanese encephalitis virus strain Nakayama (EF571853) was used as the outgroup. (B) WNV strains of cluster 2 that are most related to the dromedary WNV and strains of the other clusters within lineage 1a are indicated. A total of 10 302 nucleotide positions were included in the analysis. Kunjin strain MRM61C (D00246) of lineage 1b was used as the outgroup. The scale bar indicates the number of nucleotide substitutions per site. All names and accession numbers are given as cited in the ViPR sequence and GenBank databases. Bootstrap values are shown only for nodes that were well supported by the maximum-likelihood method (⩾700 bootstrap support).