Figures & data
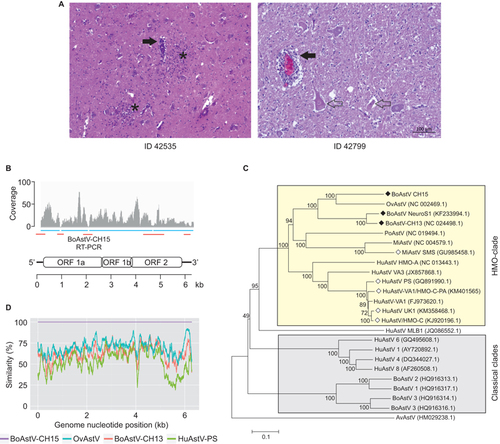
Figure 1 Identification of the novel bovine astrovirus, BoAstV-CH15. (A) Representative histopathological lesions in the brains of animal 42 535 (medulla oblongata) and 42 799 (hippocampus). Filled arrows, mononuclear perivascular cuffs; asterisks, glial nodules; open arrows, neuronal necrosis. (B) Schematic of the positive strand RNA genome of BoAstV-CH15. The four contigs obtained by NGS and bioinformatics are indicated as blue bars and read coverage is presented in a coverage plot. RT-PCR and Sanger sequencing was performed to bridge the contigs and to determine the 5′ and 3′ ends (red bars). (C) Neighbor-joining tree based on full genome nucleotide sequences. BoAstV-CH15 belongs to the human-mink-ovine (HMO) astrovirus clade (yellow box). Filled rhombus, bovine neurotropic strains; open rhombus, neurotropic strains of other species. AvAstV, avian nephritis astrovirus; HuAstV, human astrovirus; MiAstV, mink astrovirus; OvAstV, ovine astrovirus; PoAstV, porcine astrovirus. GenBank accession numbers are noted in brackets. (D) Pairwise genome comparison between selected encephalitis-associated astrovirus isolates.