Figures & data
Figure 2 ZIKV and DENV E proteins share considerable sequence identity. (A) Phylogenetic tree, showing relatedness based on E protein sequence, created using MEGA7.Citation67 The evolutionary history between the viruses was inferred by using the maximum likelihood method based on the JTT matrix-based model.Citation68 The percentage of trees in which the associated viral sequences clustered together is shown next to the branches. Branches are drawn to scale, with lengths measured in the number of substitutions per site. (B) Heat map showing E protein sequence identity, generated with ggplot2 in R.Citation69 Sequences were aligned in Geneious version 6.1.Citation70 For (A) and (B), the ZIKV strains analyzed and their GenBank accession numbers are: PRVABC59: KU501215, MR766: AY632535, H/PF/2013: KJ776791, Yap/2007: EU545988 and SPH2015: KU321639. The DENV strains are DENV1 WestPac: U88535, DENV2 Tonga/72: AY744147.1, DENV3 Sleman/78: AY648961.1 and DENV4 Dominica/814669/1981: AF326573.1. In addition, the YFV strain Asibi: KF769016.1 was also included as an outgroup in the sequence analyses above.
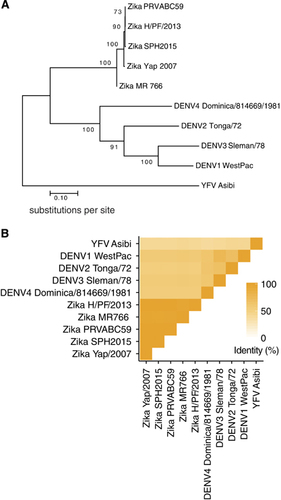
Figure 1 The ZIKV particle and E protein dimer. Cryo-EM surface structures of (A) immature (PDB 5U4W)Citation31 and (B) mature (PDB 5IRE)Citation30 ZIKV. The E protein dimer is highlighted in a yellow box. (C) The ZIKV E protein dimer colored by its domain, EDI: red, EDII: yellow and EDIII: blue.Citation30 The fusion loop is circled in orange. All structural figures in (A–C) were created using PyMol (Schrödinger LLC).
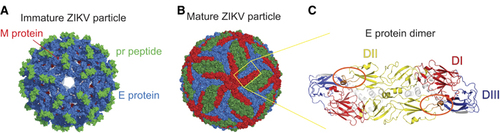
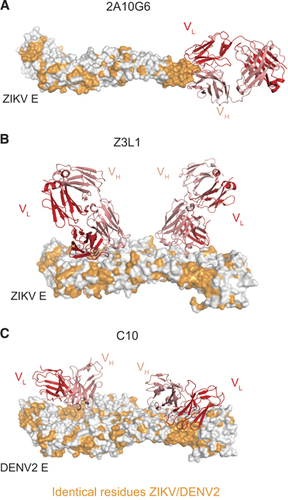
Figure 3 Cross-reactive and ZIKV-specific mAbs binding their E protein epitopes. (A) Murine mAb 2A10G6 binding to the conserved fusion loop on a ZIKV E monomer.Citation35 (B) Human mAb Z3L1 binding to a ZIKV-specific EDI epitope.Citation45 (C) Human mAb C10 (ribbon structure) binding to a dimer-dependent epitope on E protein dimer.Citation71 For (A–C), heavy and light chains of mAbs are colored red and pink, respectively. All residues conserved between DENV2 and ZIKV are colored orange. All figures were created using PyMol (Schrödinger LLC).