Figures & data
Figure 1 Avian influenza A viruses isolated from (A) the Tanguar haor and (B) live poultry markets (LPMs) of Bangladesh. (A) In February 2015, December 2015 and February 2016, 120, 1000, and 1011 samples, respectively, were collected from ducks (Khaki Campbell) from free-range farms and wild birds in the Tanguar haor area. No viruses were isolated from the samples collected in February 2016. All viruses were isolated from ducks, except one H7N5 virus, which was isolated from a wild black-tailed godwit. (B) Monthly virus isolation from LPMs in Bangladesh. From February 2015 through February 2016, 160 samples (95, 95 and 93 samples were collected in February, March and April 2015, respectively) were collected per month from five LPMs in Bangladesh. Samples collected in May, October and November 2015 did not result in any virus isolates. (C) Viruses isolated from various species of birds in LPMs. Only 18 samples were collected from geese (April 2015) during the surveillance period, from which one H5N1 virus was isolated. Virus isolation rates (× 100 number of viruses isolated/number of samples inoculated in eggs) are shown on top of each column representing different species.
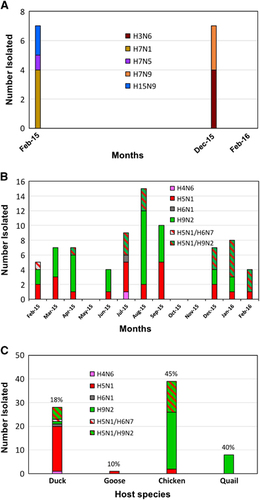
Figure 2 Phylogenetic relationship of the hemagglutinin (HA) genes of (A) H3, (B) H4, (C) H6, (D) H7 and (E) H15 low pathogenic avian influenza (LPAI) viruses isolated in Bangladesh. Viruses identified during the surveillance period are depicted in blue. •, LPAI viruses previously isolated from Bangladesh. Trees are rooted to midpoint. Bootstrap values ⩾70 are indicated on branches.
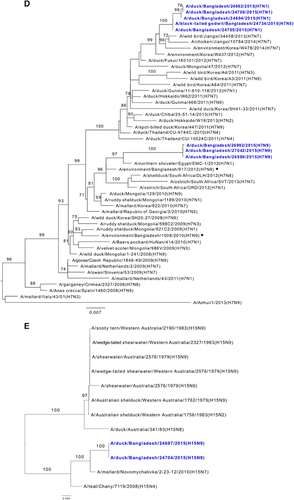
Figure 3 Phylogenetic relationship of the hemagglutinin (HA) genes of the highly pathogenic avian influenza (HPAI) H5N1 viruses isolated from Bangladeshi live poultry markets. The complete coding region of HA1 was used. The viruses identified during the surveillance period are color coded (red and purple for old and new genotype H5N1 viruses, respectively). ♦, viruses isolated in 2013 from Bangladesh with a gene constellation similar to that of new genotype H5N1 viruses described here. *, HPAI H5N1 viruses isolated from humans in Bangladesh. ⇐, reference antigens. Tree is rooted to midpoint. Bootstrap values ⩾70 are indicated on branches.
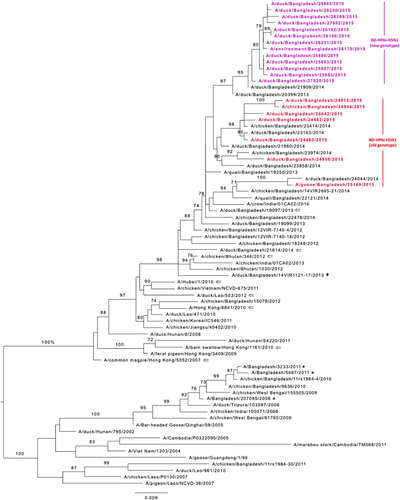
Figure 4 Phylogenetic relationship of the polymerase basic 1 (PB1) genes of the viruses isolated in Bangladesh (BD). The complete coding region, except for first 11 amino acids (2100 nucleotides total), was used. The viruses identified during the surveillance period are color coded (red, old genotype H5N1; purple, new genotype H5N1; green, H9N2; and blue, non-H9N2 LPAI viruses). •, non-H9N2 LPAI viruses previously isolated from Bangladesh. ♦, H5N1 viruses isolated in 2013 from Bangladesh with a gene constellation similar to that of new genotype H5N1 viruses described here. *, HPAI H5N1 viruses isolated from humans in Bangladesh. Tree is rooted to PB1 sequence of A/equine/Prague/1/1956(H7N7). Bootstrap values ⩾70 are indicated on branches.
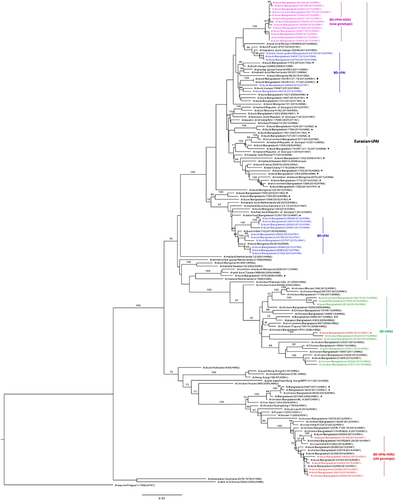
Figure 5 Gene constellations of highly pathogenic avian influenza (HPAI) H5N1 viruses isolated in Bangladesh from February 2015 through February 2016. Gene segments of circulating Bangladeshi HPAI H5N1 (red), Eurasian-lineage (blue), and H9N2-like (green) viruses are depicted for each isolated virus. Note that a newly emerged genotype of H5N1 viruses was first isolated from a single LPM in June 2015. This genotype was then isolated from other markets and became predominant in all Bangladeshi LPMs.
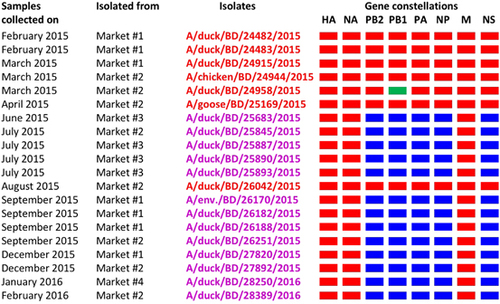
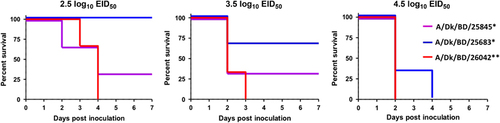
Figure 6 Survival of chickens after inoculation with newly emerged Bangladeshi highly pathogenic avian influenza (HPAI) H5N1 viruses. *, new genotype H5N1 viruses. **, old genotype H5N1 viruses circulating in Bangladesh since 2011. The inoculation doses for the A/Dk/BD/25845 and A/Dk/BD/26042 viruses were 2.5, 3.5 and 4.5 log10 EID50. The inoculation doses of the A/Dk/BD/25683 virus were 2.67, 3.67 and 4.67 log10 EID50. Duck, Dk; Bangladesh, BD.