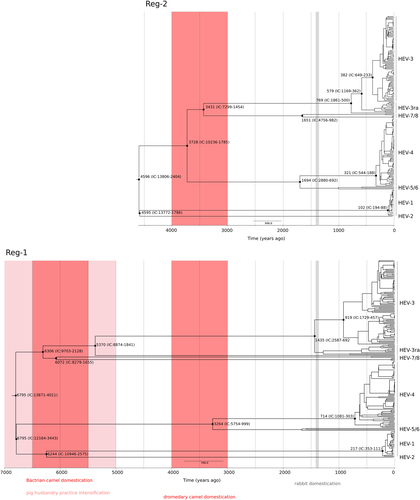
Figures & data
a Geographic distribution of the four major HEV genotypes (HEV-1 to HEV-4). b Geographic origin of complete Orthohepevirus A genomes analyzed in this study. The number of sequences deriving from each continent or region is reported in parentheses. The location where animal-infecting Orthohepevirus A were collected is also shown. c Schematic representation of the Orthohepevirus A genome. Positions refer to the Burma (GenBank accession: M73218) reference strain for genotype 1. Note that ORF4 has been described only in HEV-1 and not in other HEV genotypesCitation25. The regions we used in different analyses, as detailed in the text, are shown. The region we filtered from the Orthohepevirus A genome alignment is indicated. The location of the recombination breakpoint is also reported. MT methyltransferase, P papain-like cysteine protease, Hel helicase, RdRp RNA-dependent RNA polymerase, Y Y domain, HVR hypervariable region, also known as polyproline region (PPR); X, macro domain
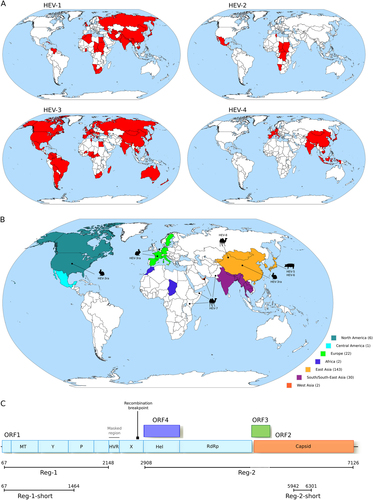
a Maximum clade credibility tree for Reg-1. Branches are colored according to inferred ancestral location; posterior support for inferred locations at relevant nodes is shown. b The topologies of the Reg-1 and Reg-2 trees are shown. Pie charts represent the ancestral location probability of the respective area. Probabilities were averaged over 100 BBM runs with a similar number of sequences from East Asia, South/South-East Asia, and Europe. In BBM analyses, combined regions appear because we allowed two areas per node. c Posterior probability distributions for the location of the Orthohepevirus A MRCA obtained using BEAST and BBM. Results refer to the Reg-2-short region
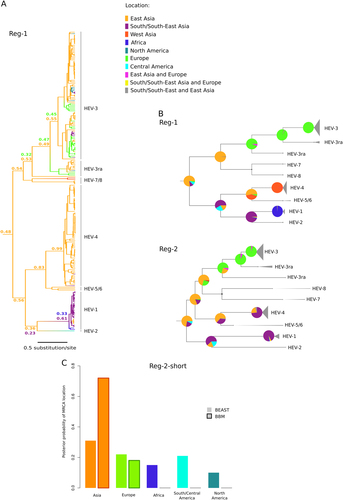
a Maximum clade credibility trees for Reg-1 and Reg-2. Branches are colored according to inferred ancestral host; posterior support for the host range of the MRCA is reported. b Posterior probability distributions of ancestral host state for the MRCA of the Orthohepevirus A phylogeny. Probabilities derive from 100 BBM runs with omission of character state information for 22 randomly sampled human sequences belonging to genotypes 3 and 4
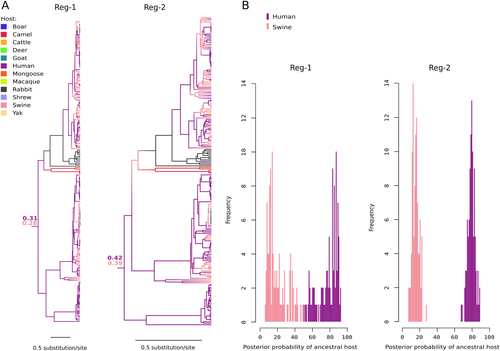
Branch lengths and tMRCA estimates for the Orthohepevirus genus
Timescaled phylogenetic tree estimated using the Reg-1 and Reg-2 regions are shown. Branch lengths represent the evolutionary time measured by the grids corresponding to the timescale shown at the tree base (in years). The tMRCA of selected nodes is reported with 95% confidence intervals. The time-frames of historical events mentioned in the text are reported