Figures & data
Detection rates of rodent bocavirus among different rodent species in Dalian, Dandong, Huludao, Manzhouli, Zhangmu, Hami, Pingxiang, Ruili and Hekou from 2011 to 2017
Detection rates of rodent bocavirus in different months in in Dalian, Dandong, Huludao, Manzhouli, Zhangmu, Hami, Pingxiang, Ruili and Hekou from 2011 to 2017
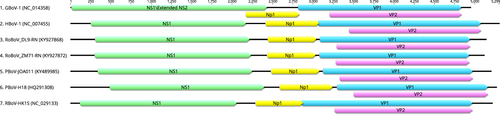
Nine isolates of rodent bocavirus identified in this study
Different colored boxes represent four ORFs of bocaparvoviruses. The illustration was constructed by geneious software. (Biomatters Ltd, Auckland, New Zealand)
Amino acid and nucleotide (data in parentheses) identities of NS1, NP1, VP1 between RoBoV isolates DL9-RN, ZM71-RN and other bocaparvoviruses
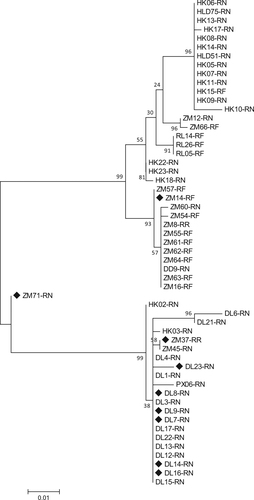
Maximum likelihood method was used to generate the tree under the best model (LG + G + I), which was selected based on the BIC scores (Supplementary Table S2). Scale bar indicates amino acid substitutions per site. Bootstrap values obtained in 100 replicates are indicated at the nodes. The sequences obtained in this study are marked with a black diamond. The tree is midpoint rooted. Isolate’s name DL8-RN denotes that the isolate’s host is Rattus norvegicus which was captured in Dalian. Likewise, the name ZM14-RF denotes this isolate’s host is Rattus flavipectus which was captured in Zhangmu. Csl bocavirus California sea lion bocavirus, GBoV gorilla bocavirus, HBoV human bocavirus, PBoV porcine bocavirus, RoBoV rodent bocavirus, RBoV rat bocavirus
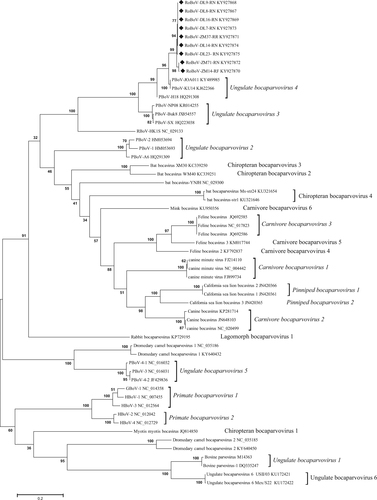
The maximum likelihood tree was constructed under the best model (HKY + G), which was selected based on the BIC scores (Supplementary Table S3). Scale bar indicates nucleotide substitutions per site. Bootstrap values obtained in 100 replicates are indicated at the nodes. The nine isolates with genome or CDS sequenced are marked with a black diamond. The tree is midpoint rooted. DL, DD, HLD, ZM, HM, MZL, PX, RL and HK in isolates’ name represent samples collected from Dalian, Dandong, Huludao, Zhangmu, Hami, Manzhouli, Pingxiang, Ruili, and Hekou respectively. Suffixes of isolates’ name denote the host species: AA Apodemus agrarius, CB Cricetulus barabensis, MM Mus musculus, RN Rattus norvegicus, RF Rattus flavipectus, RO Rhombomys opimus, RR Rattus rattus