Figures & data
Origin, China, and Korea/USA clades are represented in green, red, and orange, respectively
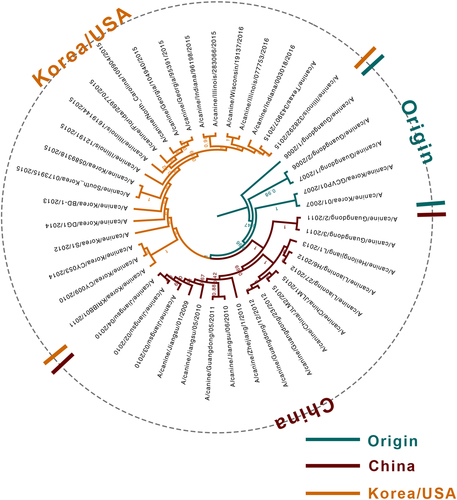
The H3N2 CIV strains are clustered into three genotypic groups, namely, Origin, China, and Korea/USA, represented in green, red, and orange, respectively
RSCU of H3N2 CIVs, genotypes, and potential hosts
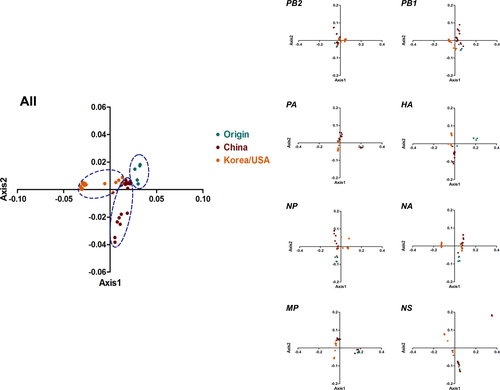
Genotype-specific PCA was structured for whole genomes and H3N2 (eight segments). Origin, China, and Korea/USA clades are represented in green, red, and orange, respectively
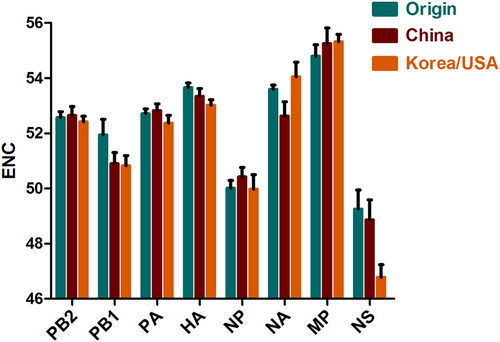
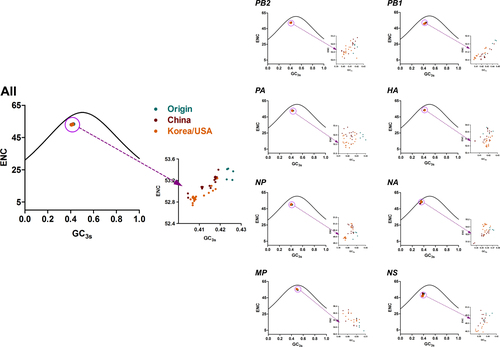
Solid curves represent the stranded ENC. Origin, China, and Korea/USA clades are represented as green, red, and orange diamonds, respectively
Correlation analysis among nucleotide composition, codon composition, ENC, Aroma, Gravy, Axis1, and Axis2
.
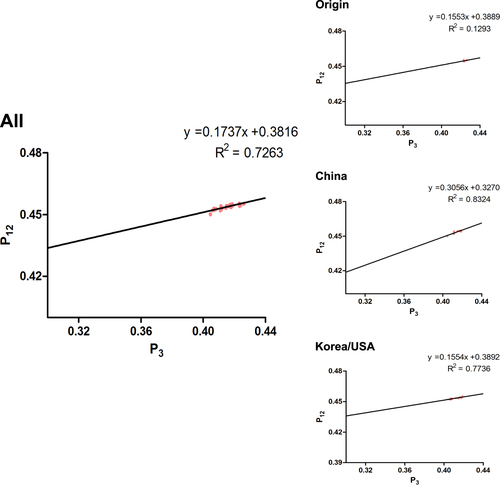
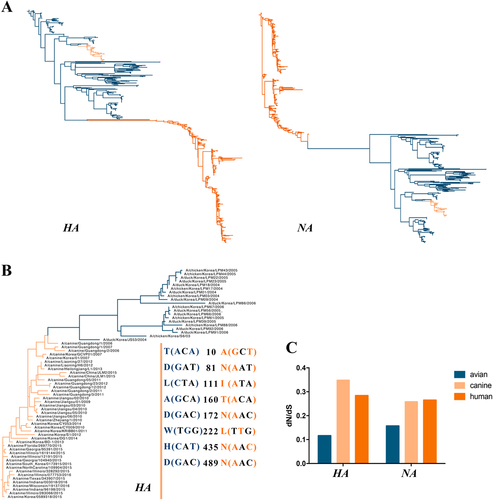
Fig. 6 ML phylogenetic trees of the HA and NA(A) genes reconstructed with 597 sequences and phylogenetic tree of the HA(B) gene constructed with 43 H3N2 CIVs and 20 AIVs and the dN/dS(C) value of the influenza viruses (including avian, canine, and human) corresponding to the HA and NA genes (H3N2 AIVs are blue, CIVs are pink, and the human influenza virus is orange)