Figures & data
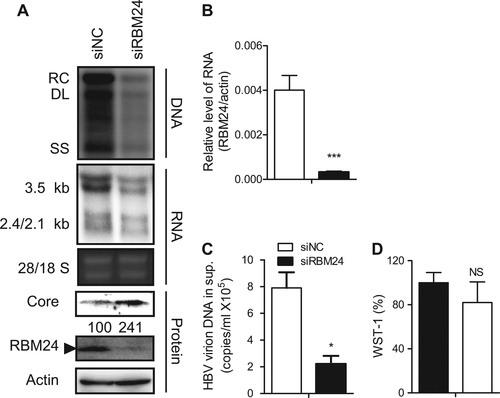
HepG2.2.15 cells were transfected with the indicated siRNA. a HBV replication intermediates were detected by southern blotting. The positions of relaxed circular (RC), double-stranded linear (DL), and single-stranded (SS) DNA are indicated (top panel). HBV transcripts were detected by northern blotting. Ribosomal RNA (28S and 18S) are presented as loading controls. The positions of HBV 3.5-kb, 2.4-kb, and 2.1-kb RNA are indicated (middle panel). RBM24 and core were detected by western blotting using an anti-RBM24 antibody or an anti-core antibody. The levels of β-actin served as a loading control (bottom panel). b The relative level of RBM24 was detected by real-time PCR. c HBV virions isolated from supernatants were used to quantify HBV DNA. d The WST-1 was detected to indicate the cytotoxic activities
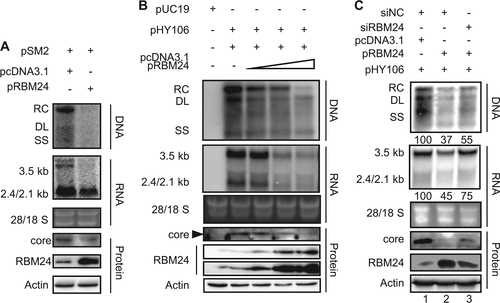
a HepG2 cell were co-transfected with 1.5 μg of pSM2 and 0.5 μg of pRBM24 or empty vector in 6-well plates. b HepG2 cells were co-transfected with 0.8 μg of pHY106 and 0, 0.01, 0.1, or 0.5 μg of pRBM24 or empty vector in 6-well plates. c HepG2 cells were co-transfected with 0.8 μg of pHY106, 0.5 μg of pRBM24, and the indicated siRNA or empty vector in 6-well plates. a–c HBV replication intermediates were detected by southern blotting. The positions of relaxed circular (RC), double-stranded linear (DL), and single-stranded (SS) DNA are indicated (top panel). HBV transcripts were detected by northern blotting. Ribosomal RNA (28S and 18S) are presented as loading controls. The positions of the HBV 3.5-kb, 2.4-kb, and 2.1-kb RNA are indicated (middle panel). RBM24 and core were detected by western blotting using an anti-RBM24 antibody or an anti-core antibody. The levels of β-actin served as a loading control (bottom panel). Hybridization signals were quantified with NIH ImageJ software
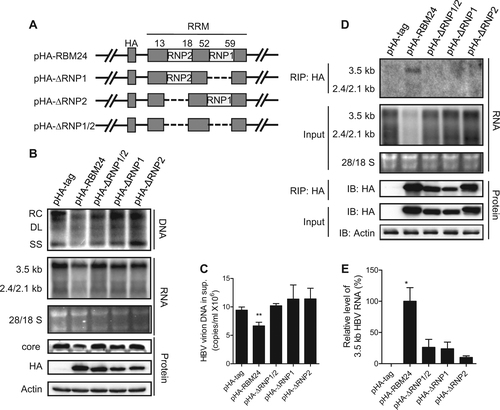
a Schematic illustration of the construction strategy of RBM24 deletion clones. b, c HepG2 cells were co-transfected with 0.8 μg of pHY106 and 0.5 μg of pHA-RBM24, pHA-ΔRNP1/2, pHA-ΔRNP1, pHA-ΔRNP2, or empty vector in 6-well plates. b HBV replication intermediates were detected by southern blotting. HBV transcripts were detected by northern blotting. RBM24 and core were detected by western blotting. c HBV virions isolated from supernatants were used to quantify HBV DNA. d, e HEK293T cells were co-transfected with 10 μg of pHY106 and 8 μg of pHA-RBM24, pHA-ΔRNP1/2, pHA-ΔRNP1, pHA-ΔRNP2, or empty vector in 100-mm dishes. Cell lysates were collected at 48 h post-transfection. RNA-IP assays were performed using anti-HA antibody. d The input or co-immunoprecipitated RNA and protein were detected. e The co-immunoprecipitated HBV 3.5-kb RNA was detected by real-time PCR
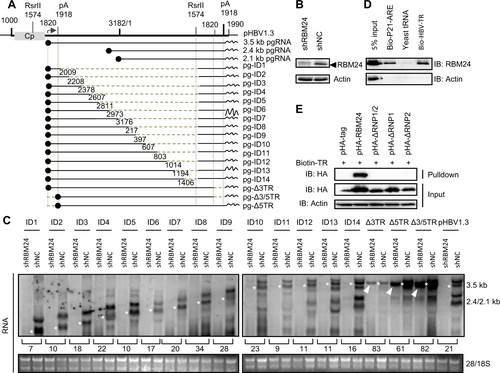
a Schematic illustration of the construction strategy of HBV deletion clones. b The level of RBM24 in the HepG2-shNC cells and HepG2-shRBM24 cells was detected by western blotting. c HepG2-shNC cells and HepG2-shRBM24 cells were transfected with HBV deletion clones. The HBV mRNA transcribed from the internal deletion clones was detected by northern blotting. The full-length or truncated pgRNA are indicated by “*”. d RBM24 cell lysates were pulled down with biotin-HBV-TR, biotin-p21-ARE (positive control), or yeast tRNA (negative control). e pHA-tag, pHA-RBM24, pHA-ΔRNP1/2, pHA-ΔRNP1, or pHA-ΔRNP2 cell lysates were pulled down with biotin-HBV-TR, and the input actin, HA-tag, HA-RBM24, HA-ΔRNP1/2, HA-ΔRNP1, and HA-ΔRNP2 served as controls
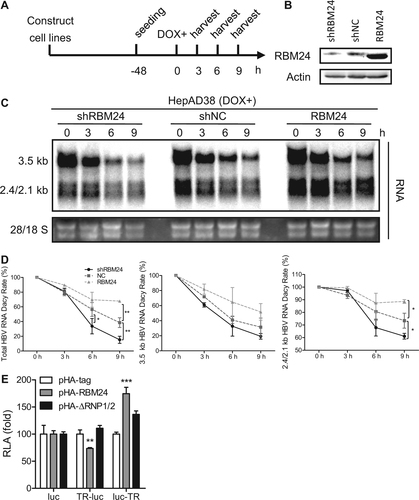
a HepAD38-shRBM24, HepAD38-shNC, and HepAD38-RBM24 cells were constructed and seeded at −48 h. DOX was added to the culture medium to shut down HBV RNA transcription at 0 h, and cells were harvested at the indicated time points. b The expression of RBM24 in cell lines was detected by western blotting. c HBV RNA was extracted from harvested cells and analyzed by northern blotting. d Kinetic analysis of HBV RNA decay in the cell lines. The relative levels of HBV RNA were normalized to 28S and expressed as the percentage of the RNA signals from the corresponding sample at time point 0 h. e HepG2 cells were co-transfected with each indicated reporter plasmid (pluc, pTR-luc, or pluc-TR) and control vector or plasmid expressing RBM24 or ΔRNP1/2. Cells were harvested at 48 hpt, and the relative luciferase activity (RLA) was measured
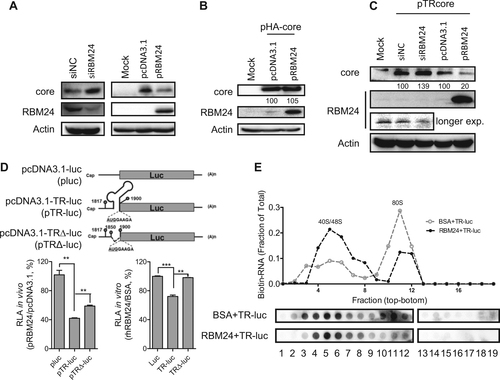
a The HepG2.2.15 cell line was transfected with siNC or siRBM24 (left panels), HepG2 cells were co-transfected with pHY106 and pRBM24 or empty vector (right panels). Cell lysates were collected at 48 hpt, and the expression of core and RBM24 was detected by western blotting. b HepG2 cells were co-transfected with pHA-core and pRBM24 or empty vector, and the expression of core and RBM24 was detected by western blotting. c HepG2 cells were co-transfected with pTR-core and the indicated siRNA or plasmid and harvested at 48 hpt. The expression of core and RBM24 was detected by western blotting. d HepG2 cells were transfected with the indicated plasmids, and luciferase activity was determined with Steady-Glo®. The relative luciferase activity (RLA) values were calculated and are shown in the bar graph on the left. The luciferase activity of 5′ TR-associated luciferase reporter plasmids in vitro was detected (bar graph on the right). e The 5′ TR-luciferase RNA together with rhRBM24 or BSA was incubated in rabbit reticulocyte lysate (RRL). The ribosome complexes were separated by sucrose gradient ultracentrifugation. The distribution of biotin-RNA was detected using a dot-blot assay
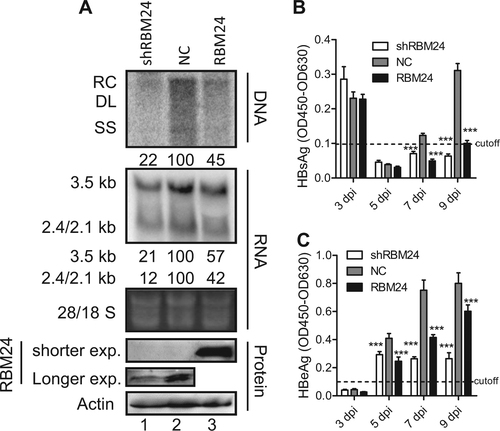
Huh7-NTCP shNC, shRBM24, and RBM24 cells were seeded into 6-well plates and spinoculated with HBV virion particles. a HBV replication intermediates were detected by southern blotting. HBV transcripts were detected by northern blotting. The 28S and 18S fragments are presented as loading controls. RBM24 were detected by western blotting. Hybridization signals were quantified using the NIH ImageJ software. b, c HBsAg and HBeAg were detected by ELISA
Oligonucleotides used in this study
