Figures & data
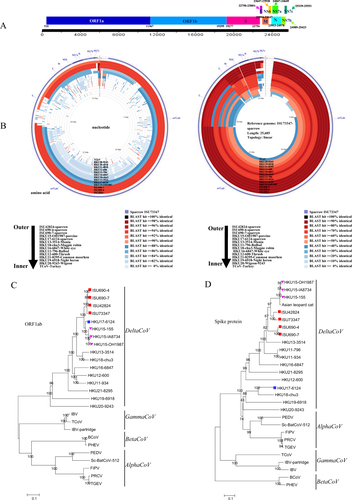
a Genome organization of SpDCoV ISU73347. b Nucleotide and amino acid comparison of ISU73347 with three sparrow coronaviruses and other reference strains (PDCoV strain HKU15-OH1987, KJ462462; sparrow CoV strain HKU17-6124, NC_016992; munia CoV strain HKU13-3514, NC_011550; magpie robin CoV strain HKU18-chu3, NC_016993; white-eye CoV HKU16-6847, JQ065044; bulbul CoV strain HKU11-796, FJ376620; thrush CoV strain HKU12-600, NC_011549; common moorhen CoV strain HKU21-8295, NC_016996; night heron CoV strain HKU19-6918, NC_016994; wigeon CoV strain HKU20-9243, JQ065048; TCoV, turkey coronavirus, NC_010800). Analysis was completed using CGView Comparison Tool software. The corresponding strains/samples for rings are detailed on the bottom. ORF open reading frame, S spike, E envelope, M membrane, N nucleocapsid, NS6, NS7a, NS7b, and NS7c represent nonstructural proteins 6, 7a, 7b, and 7c, respectively. c Phylogeny of ORF1ab amino-acid sequences. d Phylogeny of spike protein sequences. The sparrow deltacoronavirus strains identified in this study are indicated with red squares, HKU17-6124 is marked with a blue square, and three HKU15 strains are marked with a pink triangle. The phylogeny was inferred using the neighbor-joining method in MEGA version 7.0. Statistical support was obtained using bootstrap resampling (1000 replications) and is drawn on the inferred tree. Reference sequences representing coronavirus diversity were obtained from NCBI GenBank (PEDV, porcine epidemic diarrhea virus, NC_003436; Sc-BatCoV-512, Scotophilus bat coronavirus 512, NC_009657, TGEV transmissible gastroenteritis virus, AJ271965, FIPV feline infectious peritonitis virus, NC_002306, PRCV porcine respiratory coronavirus, DQ811787, PHEV porcine hemagglutinating encephalomyelitis virus, DQ011855, BCoV bovine coronavirus, NC_003045, IBV infectious bronchitis virus, NC_001451, IBV-partridge partridge coronavirus, AY646283, TCoV turkey coronavirus, NC_010800; bulbul, CoV strain HKU11-796, FJ376620; bulbul CoV strain HKU11-934, FJ376619; thrush CoV strain HKU12-600, NC_011549; munia CoV strain HKU13-3514, NC_011550; PDCoV strain HKU15-OH1987, KJ462462; PDCoV strain HKU15-IA8734, KJ567050; PDCoV strain HKU15-155, JQ065043; white-eye CoV strain HKU16-6847, JQ065044; sparrow CoV strain HKU17-6124, NC_016992; magpie robin CoV strain HKU18-chu3, NC_016993; night heron CoV strain HKU19-6918, NC_016994; wigeon CoV strain HKU20-9243, JQ065048; common moorhen CoV strain HKU21-8295, NC_016996). The scale bar represents 0.1 amino acid substitutions per site