Figures & data
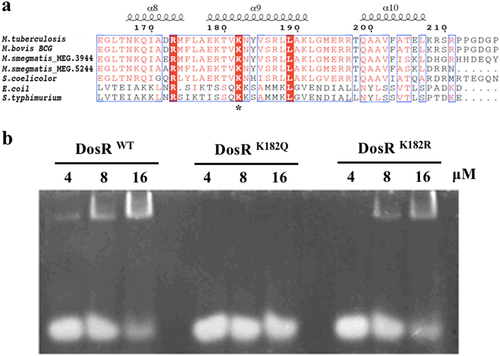
a Conservation analysis of the DosR sequence surrounding K182 from various bacteria, including Mycobacterium, Streptomyces, Escherichia, and Salmonella strains. Asterisk indicates the conserved lysine residues. b Electrophoretic mobility shift assay (EMSA) analysis using DosR and derivatives and a consensus DosR regulon 20-bp DNA fragmentCitation8. EMSA was used to evaluate the DNA-binding abilities of DosR and its derivatives at three concentrations (4 μM, 8 μM, and 16 μM). EMSA results are representative of three independent experiments
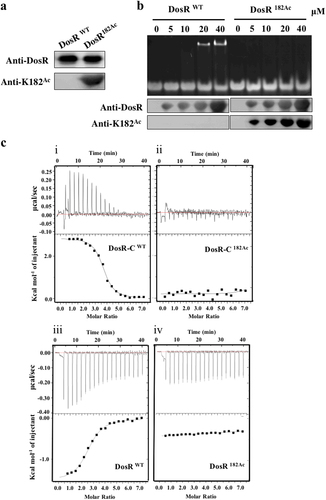
a Western blot analysis of purified wild-type DosR (DosRwt) or DosR protein in which K182 was acetylated (DosR182Ac). Antibodies specific for DosR (anti-DosR, 1:10,000) and acetylated K182 peptides (anti-K182Ac, 1:1000) were used, and the exposure time was 30 s (for anti-K182Ac) or 3 s (for anti-DosR). b DNA-binding abilities of DosR and DosRK182Ac. Electrophoretic mobility shift assay (EMSA) was used to evaluate the DNA-binding abilities of DosR and its derivatives at the indicated concentrations (lanes 2–5 and lanes 7–10) to a 20-bp DNA fragment. The acetylation level of DosRK182Ac in EMSA was tested using western blot. c Binding of full-length DosR or DosRK182Ac and the C-terminal domain of DosR (DosR-C, residues 144–217) or DosR-CK182Ac to DNA by isothermal titration calorimetry (ITC). The representative raw ITC data and the fitted binding curves are shown for the proteins DosR (i), DosRK182Ac (ii), DosR-C (iii), and DosR-CK182Ac (iv). Results are representative of at least two independent experiments
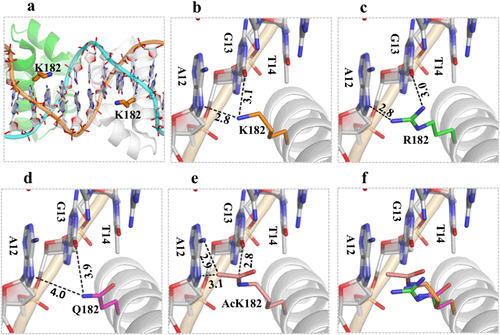
The structure of DosR was modeled using the crystal structure (PDB_ID: 1ZLK) of M. tuberculosis DosR-C in complex with DNA as its template. a The data show how K182 is located in the middle of helix 9, and its side chain points toward the nucleotide bases of DNA from the major-groove side. b Two H-bond interactions between K182 and A12 or K182 and G13 play the most important role in DNA recognition. c The K182R mutant can form two H-bonds with the target DNA to maintain its DNA-binding ability. d The K182Q mutant increases the distance between K182 and A12 or K182 and G13 and weakens the interaction with the target DNA. e The acetylated K182 has close contact between the side chain of the acetylated K and the DNA nucleotide bases. The superimposition shows the change in distance between the side chains at position 182 for different residues and the target DNA (f)
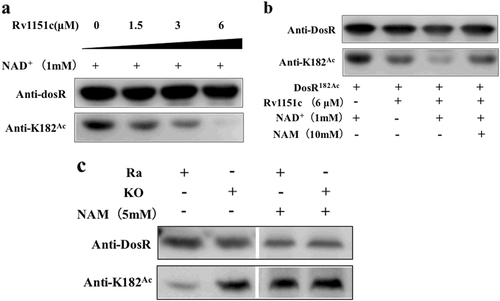
a Rv1151c deacetylates DosR in a dose-dependent manner in vitro. Purified DosR182Ac (8 µM) was incubated with NAD+ (1 mM) and Rv1151c at the indicated concentrations (1.5 µM, 3 µM, and 6 µM) at 25 °C for 2 h. The acetylation levels were evaluated by western blot with anti-DosR (1:10,000) at 3-s exposure and anti-K182Ac (1:1000) at 30-s exposure. The results are representative of three independent experiments. b Deacetylation of DosR is dependent on NAD+ and is inhibited by NAM. Purified DosR182Ac (8 µM) was incubated with or without Rv1151c (6 µM), NAD+ (1 mM), and NAM (10 mM) at 25 °C for 2 h. The acetylation levels were evaluated by western blotting with anti-DosR (1:10,000) at 3-s exposure and anti-K182Ac (1:1000) at 30-s exposure. Results are representative of three independent experiments. c The acetylation levels of DosR in H37Ra wild-type (Ra) and MRA_1161 (Rv1151c, homolog in H37Ra)-deletion mutant (KO) in vivo. H37Ra and KO were inoculated into 7H9–10% OADC–0.05% Tween 80 medium and grown to mid-log phase (OD600 = 0.4–0.6), respectively. The cultures were incubated with or without NAM (5 mM) for 10 h and collected by centrifugation. Cell extracts (20 μg per lane) were analyzed by western blotting with anti-DosR (1:10,000) at 30-s exposure and anti-K182Ac (1:1000) at 10-min exposure. Equal amounts of loading control for western blotting were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie blue (Fig. S-a, in Supplementary material). Results are representative of three independent experiments
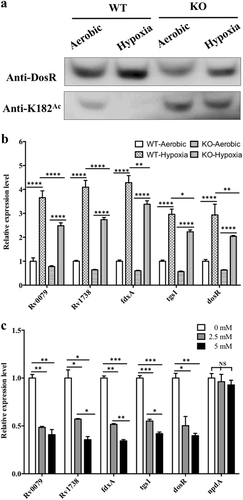
a Acetylation pattern of K182 in DosR from WT and KO mutant under aerobic or hypoxic conditions. The WT and KO mutant bacteria were cultured in Dubos medium at 37 °C for 2 days. The aerobic cultures were grown in 50-ml tubes (10 ml of culture), and hypoxic cultures were grown in 50-ml wax-sealed bottles with a headspace-to-medium ratio of 0.5. Cell extracts (20 μg per lane) were analyzed by western blotting with anti-DosR (1:10,000) at 30-s exposure and anti-K182Ac (1:1000) at 10-min exposure. Equal amounts of loading control for western blotting were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie blue (Fig. S-b, in Supplementary material). Results are representative of three independent experiments. b The transcript levels of candidate genes in the H37Ra strain and KO mutant under aerobic or hypoxic conditions. RNA was isolated from the strains grown in Dubos medium under aerobic or hypoxic conditions (see above), and the transcript levels of the candidate genes were determined by qPCR using the method of 2−ΔΔCt. The data were normalized to rrs and compared to WT. Two-way ANOVA was performed to compare WT and KO in each condition; *p < 0.05; **p < 0.01; ****p < 0.0001. c Analysis of the transcript levels of candidate genes and npdA in the H37Ra strain in the presence of NAM. H37Ra was inoculated into 7H9–10% OADC–0.05% Tween 80 medium and grown to mid-log phase (OD600 = 0.4–0.6). The cultures were incubated with NAM at different concentrations (0 mM, 2.5 mM, and 5 mM) for 10 h. RNA was isolated, and the transcript levels of candidate genes were analyzed using qPCR. The data were normalized to rrs. Statistical analysis was performed by Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001; NS: not significantly different (p > 0.05)
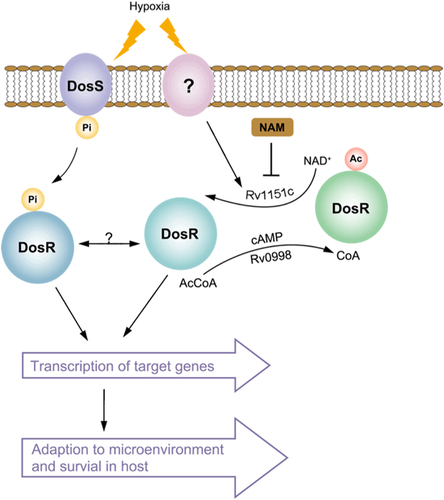
Active DosR condition: deacetylated DosR by Rv1151c enhances DNA-binding ability of DosR and then promotes the transcriptional activity of target genes, allowing M. tuberculosis to adapt to hypoxia and transition to latency in host. Inactive DosR condition: acetylated DosR by Rv0998 inhibits DNA-binding ability, which abolishes transcriptional activity of target genes. Ac acetylation, Pi phosphorylation, Ac-CoA acetyl-coenzyme A, NAD+ nicotinamide adenine dinucleotide, NAM nicotinamide
Primers used in this study
