Figures & data
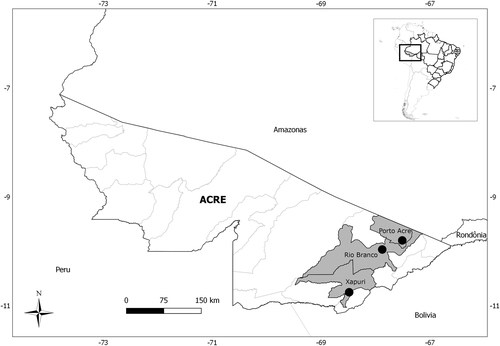
Rodents captured in three localities from Acre state, Brazil, 2015–2016, by species and locality
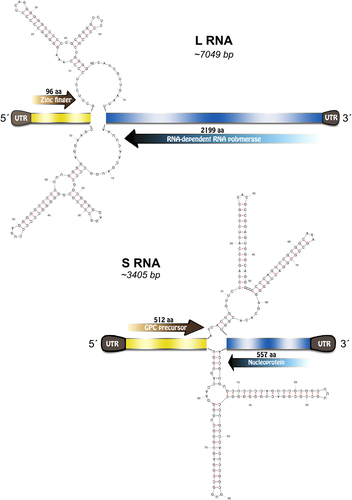
Fig. 2 Xapuri mammarenavirus genome organization and potential secondary structure of intergenic regions
Nucleotide and amino acid identities of XAPV compared with New World representatives of the genus Mammarenavirus
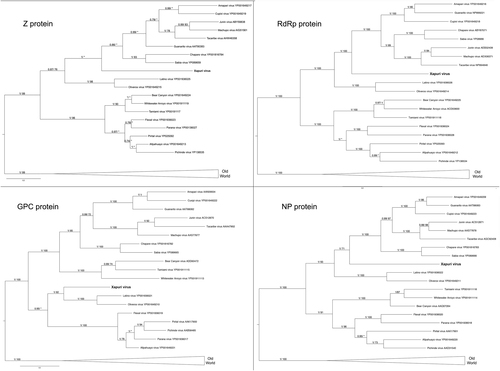
. Numbers (≥0.7/≥70) above branches indicate node probabilities or bootstrap values (MrBayes/ML). Asterisks indicate values below 0.7/70. Sequences from this study are highlighted in bold
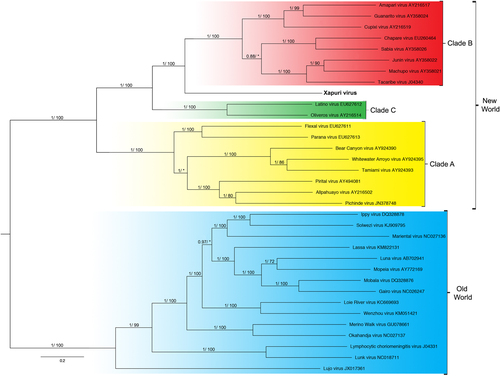
. Numbers (≥0.7/≥70) above branches indicate node probabilities or bootstrap values (MrBayes/ML). Asterisks indicate values below 0.7/70. Sequences from this study are highlighted in bold
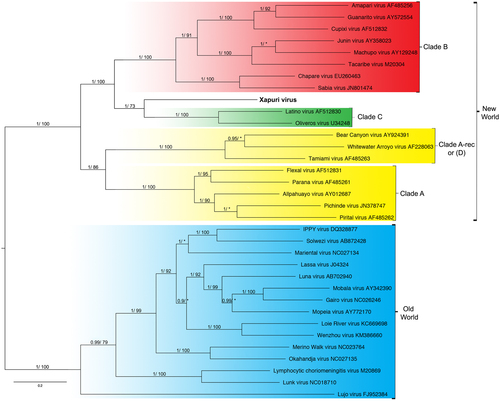
Numbers (≥0.7/≥70) above branches indicate node probabilities or bootstrap values (MrBayes/ML). Asterisks indicate values below 0.7/70. †Exhibited a difference between ML and MrBayes tree-building method topologies. Sequences from this study are highlighted in bold