Figures & data
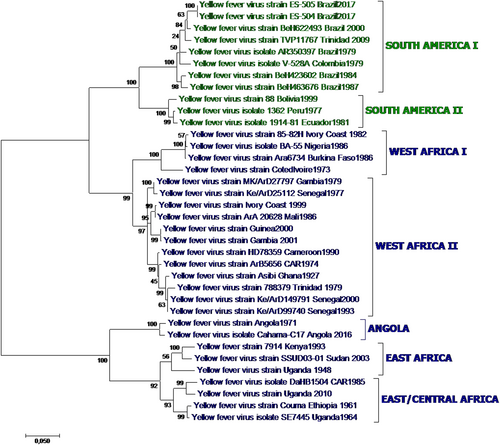
Fig. 1 Yellow fever phylogenetic analysis showing major YFV genotypes, based on alignment of a 1428 nt region of the prM-E junction region for 36 representative African and American YFV strains (Table ) using the Maximum Likelihood method based on the general time reversible model (GTR). Individual strains are defined by name and country/year of isolation. Bootstrap values (500 replicates) for major branches are indicated
Yellow fever virus strains (A: vaccine strains, B: wild type strains) included in the evaluation of suitability of published molecular detection assays
The figure is restricted to the four assays described in referencesCitation67,Citation69,Citation70,Citation75, which generated the fewest mismatches overall in the comparison with the reference set of 61 YFV genomic sequences (Table ). Perfectly matched YFV sequences are not shown. Primer and probe sequences are written 5′ to 3′ except for the reverse primers at the right edge of the figure, which are represented by the reverse-complement of the oligonucleotide sequence

The figure is restricted to the two assays described in referencesCitation69,Citation76, which generated the fewest mismatches overall in the comparison with the reference set of 61 YFV genomic sequences (Table ). Perfectly matched YFV sequences are not shown. Primer and probe sequences are written 5′ to 3′ except for the reverse primers at the right edge of the figure, which are represented by the reverse-complement of the oligonucleotide sequence

