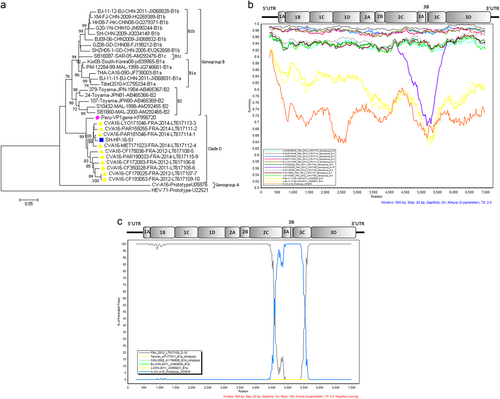
Figures & data
Fig. 1 a Phylogenetic tree based on complete CV-A16 VP1 sequences. The tree was inferred with the neighbor-joining method from genetic distances calculated using the P distance algorithm. The tree topology was assessed with 1000 bootstraps. CV-A16 strain G10 (South Africa, 1951) was the only sample assigned to clade A. HEV 71 BrCr was used as an outgroup. b Nucleotide similarity between all genogroups of CV-16 full-length genomes. The similarity plots were obtained with a dataset of 14 CV-A16 genomes, including 10 clade D strains reported in France, the CV-A16 strain G10 and two clade B strains. The Shanghai isolate SH-HP-16-51 was used as the query sequence. G10, BJ/CHN/2011(JX068828) and BJ/CHN/2011(JX068831) were chosen as the representative strains of genotype A, B1a and B1b, respectively. A sliding window of 500 nucleotides in 20 nucleotide steps was used in this analysis. c Bootscan analyses of clades A, B and D of CV-A16 strains on the basis of full-length genomes. The Shanghai isolate SH-HP-16-51 was the query sequence. G10, FRA12(LT617109), BJ/CHN/2011(JX068828) and BJ/CHN/2011(JX068831) were chosen as the reference strains of A, D, B1a and B1b, respectively. AF177911 and AY790926 were chosen as the representative intratypic recombinant strains. The vertical axis indicates the percent nucleotide identity between the parental sequences. The horizontal axis shows the nucleotide positions of the nucleotide sequences