Figures & data
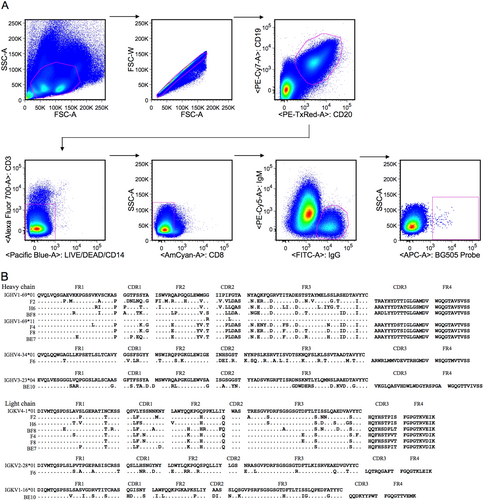
a Isolation of antigen-specific single B cells by flow cytometry. Single cells were sorted into a 96-well PCR plate containing lysis buffer according to the presented gating strategy. b Amino acid sequence analysis of monoclonal antibodies with alignment to respective germline genes. The framework region (FR) and complementarity determining region (CDR) were determined based on the program IMGT/V-QUEST. The symbol “.” denotes conserved amino acids
Gene family analysis of monoclonal antibodies
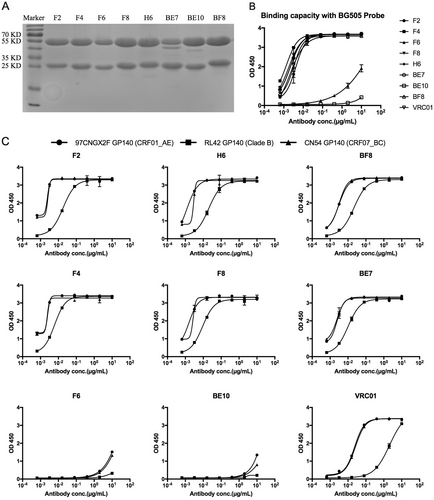
a Reducing 12% SDS-PAGE analysis of monoclonal antibodies. b ELISA binding of monoclonal antibodies with the sorting probe BG505. c ELISA of cross-reactive monoclonal antibodies binding with diverse HIV-1 GP140 antigens including clade B, CRF01_AE, and CRF07_BC
Neutralizing activity of monoclonal antibodies on the Global Panel and two Tier 1 viruses from the DRVI Panel
Neutralizing activity of F6 against diverse HIV-1 isolates on the DRVI Panel
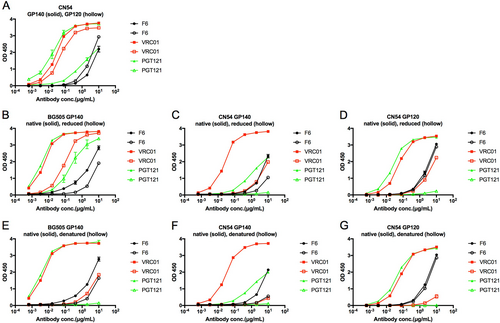
a ELISA binding of F6 to HIV-1 GP140 and GP120 antigens. b–d ELISA binding of F6 to native and reduced BG505 GP140, CN54 GP140, and CN54 GP120 treated with 10 mM DTT at 37 °C for 1 h. e–g ELISA binding of F6 to native and denatured BG505 GP140, CN54 GP140, and CN54 GP120 treated with denaturing buffer (New England Biolabs) at 100 °C for 10 min
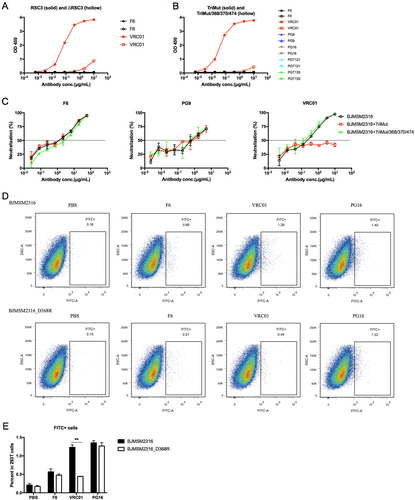
a ELISA binding of F6 to RSC3 and ΔRSC3. b ELISA binding of F6 to TriMut and TriMut/368/370/474. c Competition neutralizing of F6 against HIV-1BJMSM2316 with TriMut and TriMut/368/370/474. d Flow cytometry dot plots showing the binding capacity of F6 to HIV-1BJMSM2316 and HIV-1BJMSM2316_D368R GP160 on the surface of transfected 293 T cells. e The percentage of FITC + cells in 293 T cells are shown as mean ± SD (n = 3). The symbol “**” indicates P < 0.01
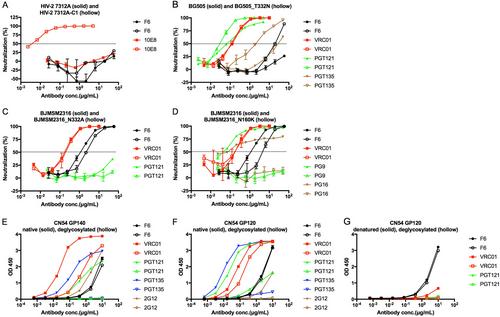
a Neutralization of F6 against HIV-27312A and HIV-27312A-C1. b Neutralization of F6 against HIV-1BG505 and HIV-1BG505_T332N. c Neutralization of F6 against HIV-1BJMSM2316 and HIV-1BJMSM2316_N332A. d Neutralization of F6 against HIV-1BJMSM2316 and HIV-1BJMSM2316_N160K. e–g ELISA binding of F6 to deglycosylated CN54 GP140 and GP120 treated with Endo H enzyme (New England Biolabs) at 37 °C according to the manufacturer’s protocol