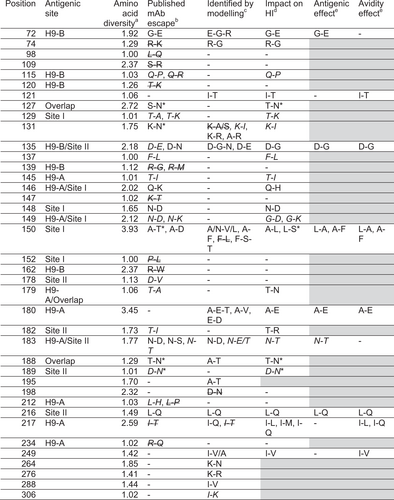
Figures & data
Change in log2 HI titre associated with stated amino-acid replacements, based on eight individual anti-UDL1/08 antisera with a minimum of five technical repeats, are indicated by cell colouring according to the colour legend with non-significant differences shown in grey and untested substitutions in white. Published mAb escape mutations are shown to the left of centre, whereas alternative substitutions are shown to the right. Mutations that introduce absent or almost absent amino-acid states ( < 1% of sequenced natural isolates) are labelled in smaller, italicised, grey font. * addition of potential glycosylation site. † would not rescue. ‡ Q146H rescued with additional substitution T186K
Frequency of potentially antigenic glycosylation sites in natural H9 isolates
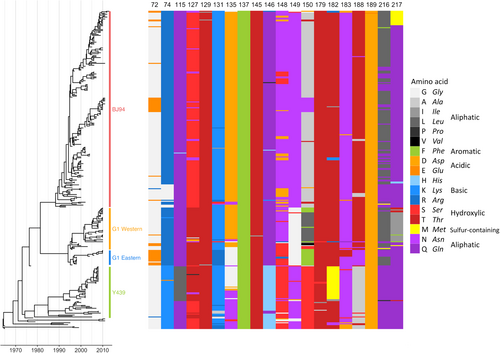
Time-resolved Bayesian HA1 phylogeny and amino-acid identity at each of the 20 mAb escape residues found to significantly affect HI titres with polyclonal chicken antisera. Amino-acid identity is shown by colour, grouped by side-chain property, according to the legend. Each virus (n = 330) included in the phylogeny has associated HI data and was included in integrated modelling of genetic and antigenic data
Amino acids substitutions correlating with antigenic change and corresponding antigenic sites
Changes in HI titre (log2) associated with each introduced substitution are shown by heatmap, measured using antiserum both lacking the introduced substitution (parent-like) and possessing the introduced mutation (mutant-like) from combinations of the viruses UDL1/08, Em/R66 and HK/33982. Changes in HI titre associated with each pair of substitutions were interpreted as effects of altered avidity and antigenicity using Equation 1, are also shown. Avidity effects cause an increase (blue) or drop (orange) in titre depending on the direction of substitution while antigenic effects are the estimated change in titre resulting from antigenic dissimilarity. X indicates not individually significant
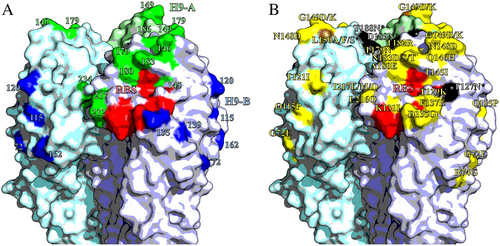
Homotrimers of H9 HA. Selected receptor-binding residues shown in red (P92, G128, T129, S130, S131, A132, W142, N173, L184, Y185, N214, G215, G218 and R219). Images made in PymolCitation52 (Schrödinger) based on the structure of A/swine/Hong Kong/9/1998 (Protein databank ID:1JSD)Citation53. a Residues recognised by mouse mAbs, positions with updated site H9-A shown in green, H9-B residues shown in blue. b Residues labelled with substitutions that affect the binding of chicken polyclonal antisera. Non-glycosylation altering substitutions shown in yellow; glycosylation site adding mutations shown in black; site 150, which had both glycosylation adding and non-glycosylation adding mutations shown in brown