Figures & data
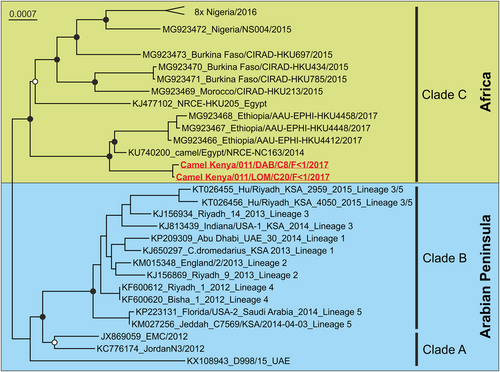
Virus designations include GenBank accession numbers and strain names. Black circles at nodes indicate bootstrap supports of >90% and white circles>75% (500 replicates). The top clade “8x Nigeria/2016” was collapsed for graphical reasons and contains eight MERS-CoV sequences (Acc. No. MG923474-81) from dromedaries in Nigeria in 2016