Figures & data
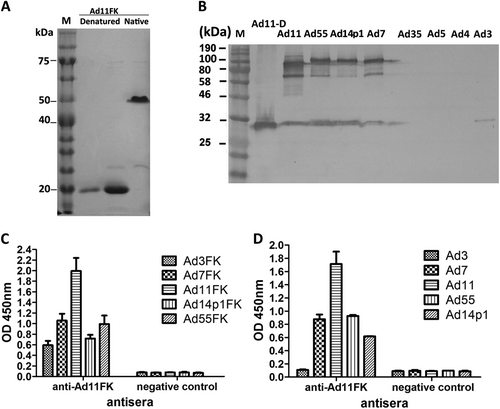
a SDS-PAGE of the purified recombinant HAdV11 fiber knob (HAdV11FK). Purified HAdV11FK was mixed with 5 × loading buffer, incubated at room temperature for 5 min, and then incubated on ice (native) or heated at 98 °C for 5 min (denatured). b Immunoblot analysis indicated that the anti-HAdV11FK serum recognizes the HAdV11 fiber in its trimeric form and cross-reacts fibers of HAdV7, HAdV14p1, and HAdV55. Purified HAdV3, HAdV4, HAdV5, HAdV7, HAdV11, HAdV14p1, HAdV35, and HAdV55 virions were stored at room temperature or heated at 98 °C (HAdV11-D) for 5 min in the presence of loading buffer. The membranes were subsequently incubated with antiserum from a mouse immunized with recombinant HAdV11FK. M, PM5000 ExcelBand™ 3-color Pre-+Stained Protein Ladder, Regular Range (SMOBIO, Taiwan (R.O.C.)). c, d Cross-reactions of anti-HAdV11FK sera with recombinant HAdV3, HAdV7, HAdV11, HAdV14p1 and HAdV55 fiber knobs (c) or purified HAdV3, HAdV7, HAdV11, HAdV14p1 and HAdV55 virions (d) were detected by ELISA. Antiserum from mice immunized with PBS was used as the negative control. Each experiment was repeated independently at least three times, and the means ± standard deviations are shown. OD450nm, optical density at 450 nm
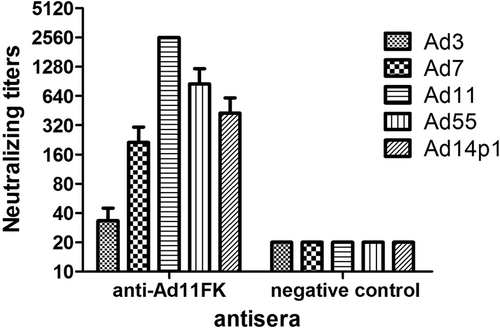
Sera from HAdV11FK-immunized mice (n = 3 per group) were assessed for NAb titers to HAdV3, HAdV7, HAdV11, HAdV55, or HAdV14p1 viruses. Antiserum from a mouse immunized with PBS was used as the negative control
Generation of Ad11-neutralizing MAbs
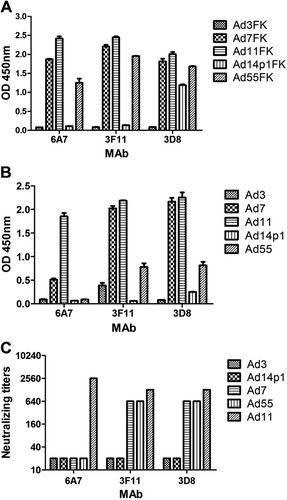
The reactions of MAbs with recombinant HAdV3, HAdV7, HAdV11, HAdV14p1, and HAdV55 fiber knobs (a) or purified HAdV3, HAdV7, HAdV11, HAdV14p1, and HAdV55 virions (b) were detected by ELISA. Each experiment was repeated independently at least three times, and the means ± standard deviations are shown. OD450nm, optical density at 450 nm. c In vitro micro-neutralization test of MAbs. The MAbs were assessed for NAb titers to HAdV3, HAdV7, HAdV11, HAdV55, or HAdV14p1 viruses
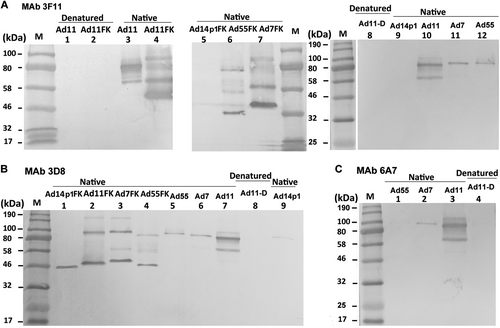
Purified HAdV7, HAdV11, HAdV14p1, and HAdV55 virions or recombinant fiber knob (FK) were stored at room temperature (“native”) or heated at 98 °C (“denatured”) for 5 min in the presence of loading buffer. The membranes were subsequently incubated with the MAbs 3F11 (a), 3D8 (b) or 6A7 (c), after which they were incubated with an HRP-conjugated secondary antibody and developed with TMB substrate. M, standard prestained protein marker (NEB, Hitchin, UK)
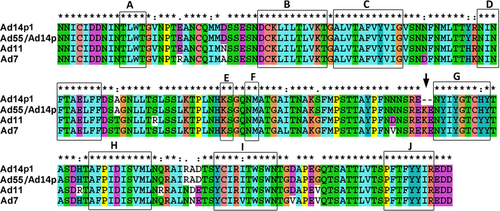
“*”, conserved amino acid; “.”, either size or hydropathy is conserved; and “:”, both size and hydropathy are conserved. Gaps used to optimize alignments are indicated by dashes. Beta sheets a–j present in the knobs are indicated by rectangles. The deletion of two amino acid residues (251KE252) within the F-G loop of the HAdV14p1 fiber knob is indicated by an arrow
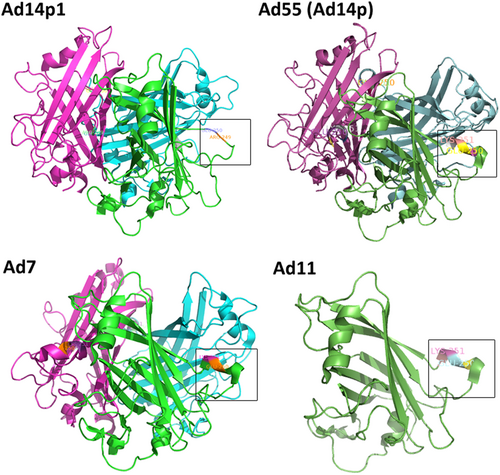
PyMOL v0.99 was used to generate the cartoon representation of the fiber knob structures. The partial residues for the F-G loop are shown in frame and are numbered according to the fiber. The EK deletion in HAdV14p1 breaks the alpha helix, as indicated
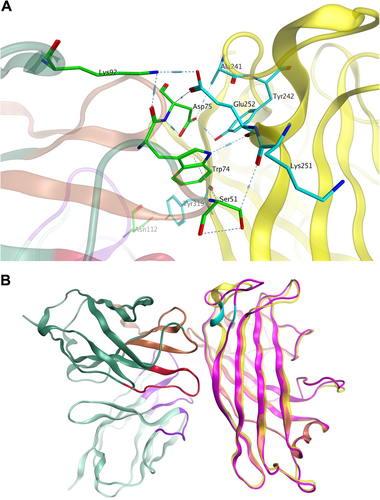
a Major binding interface between the MAb 3F11 and HAdV11FK. The residues involved in strong intermolecular contacts are shown in stick representations. The antibody residues are colored in green, and the antigen residues are colored in cyan. Strong polar interactions, such as hydrogen bonds and salt bridges, are shown in light blue dashes. b Overview of the docked complex of the antigen HAdV11FK and the antibody Fv, superimposed with HAdV14p1FK. The protein backbones are shown in ribbons. The antigen HAdV11 is colored in yellow, and HAdV14p1FK is colored in magenta. The antibody Fv is colored in dark green (FR), purple (CDR-L1/L2/L3), brown (CDR-H1/H2), and red (CDR-H3), respectively. The location of E252 on the antigen is colored in cyan