Figures & data

Figure 1. Schematic of the typical process required to compute properties of soft matter systems from system ‘chemistry’, which refers to chemical composition and state (including temperature, pressure and composition), starting from the need to either gather or derive force field parameters to model the system. For coarse-grained (CG) simulations, the CG force fields are often derived from atomistic simulations.
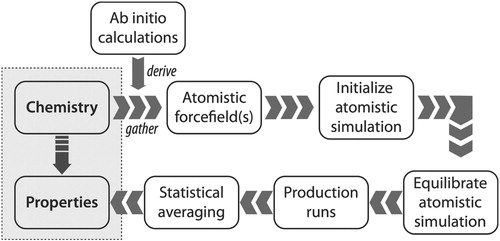
Figure 2. Python script that uses mBuild to define a class for a –CH– group, create a polymer composed of multiple –CH
– groups, and connects copies of this polymer to a surface. Note for simplicity, the terminal CH
group is not shown.
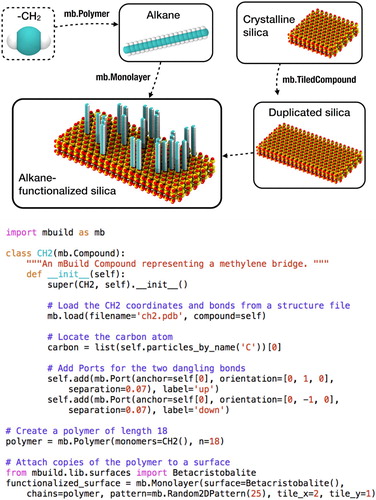
Figure 3. Two boxes of ethane constructed in mBuild. Liquid phase (left) and vapour phase (right) are simulated simultaneously in GEMC.
Figure 4. Vapor pressure (left), vapour density (middle), and liquid density (right) plots for ethane at 236 K, using GEMC in GOMC with the TraPPE force field.
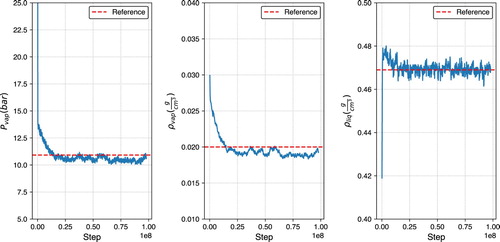
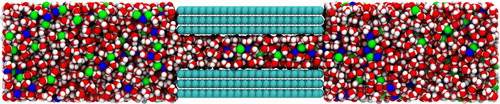
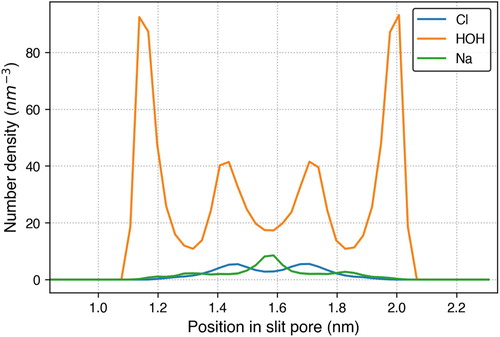
Figure 5. A snapshot of the graphene slit pore system containing graphene carbon (cyan), water (red for oxygen and white for hydrogen), sodium ions (blue) and chlorine ions (green).