Figures & data
Table 1. Primers and polymerase chain reaction protocols used in this study.
Table 2. Collected information and GenBank accession numbers for abalone species used in this study.
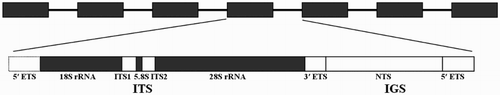
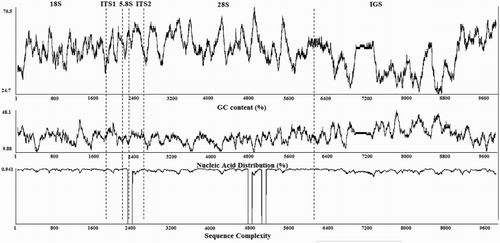
Table 3. Nucleotide composition, length, and GenBank accession number of each rRNA region from H. rubra.
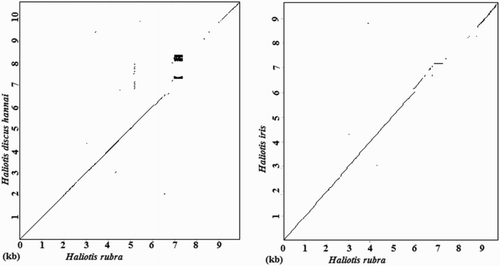
Table 4. Pairwise distances of 18S sequences (lower left matrix) and 28S rDNA sequences (upper right matrix).
Figure 2. A ML phylogenetic tree was constructed using the ‘GIR + G’ model with 1000 bootstrap replications. The numbers represent the bootstrap value (values <60% are not shown). 2n indicates the diploid chromosome number.
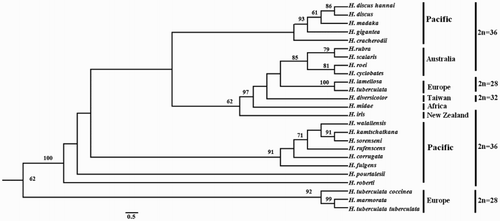
Figure 3. The entire nucleotide sequence of the H. rubra rDNA IGS region. The repeated sequences are underlined as follows: single line for repeat type A and double line for repeat type B. A putative termination signal (poly(T) tract) is in the solid box, and the RNA polymerase I transcription promoter is in the dashed box.