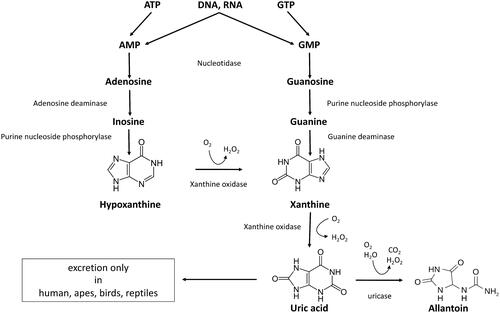
Figures & data
Table 1. Chromatographic conditions.
Table 2. Specific mass spectrometric parameters and retention times (RT) for the analytes and internal standards (IS).
Table 3. General mass spectrometric parameters.
Table 4. Concentration of labeled standards in the IS mixture and non-labeled standards in the calibrators’ mixture.
Table 5. Composition of four times concentrated AU.
Figure 2. Matrix effects. The results are quadruplicates of different dilutions of over-concentrated urine samples. Obtained concentrations were multiplied by the corresponding dilution factor and normalized to the average concentration of 20× diluted sample taken as 100% The dots and whiskers represent mean ± SD. Stars above the dots represent significant mean difference at p-levels: * < 0.05, ** < 0.001, *** < 0.0001.
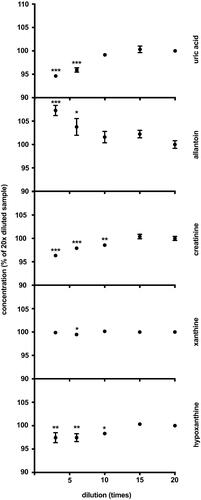
Figure 3. Storage stability. The results are quadruplicates of urine aliquots stored at different conditions for 5.5 years. The dots and whiskers represent mean ± SD. Stars above the dots represent significant mean differences at p-levels: * < 0.05, ** < 0.0003, *** < 0.0001.
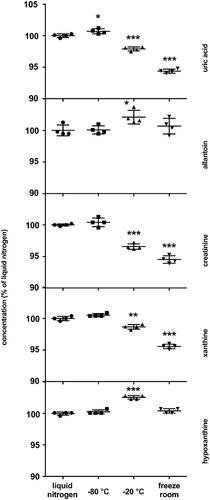
Figure 4. Typical LC–MS/MS chromatograms of four purines and creatinine in human urine. x-axis: retention time (min); y-axis: relative abundance (%).
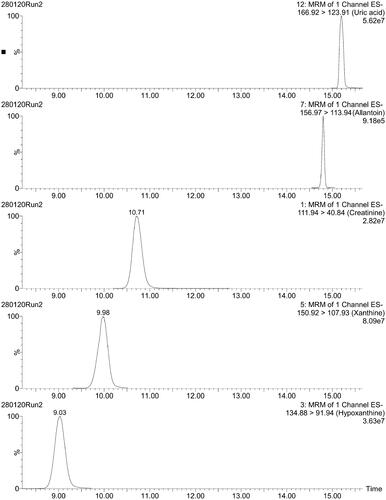
Table 6. Concentration of purine metabolites and creatinine in 1st void morning urine by LC-MS/MS.
Table 7. Inter-day variation for QC samples.
Table 8. Intra-day variation, CV%.
Figure 5. Dynamics of changes in creatinine concentration in quality controls (QC) samples. Circles: high QC; squares: low QC; dashed line: linear regression of high QC; R2 = 0.894, slope −0.454 mM/day ± 0.0639 SE; dotted line: linear regression of low QC, R2 = 0.841, slope −0.17 mM/day ± 0.033 SE.
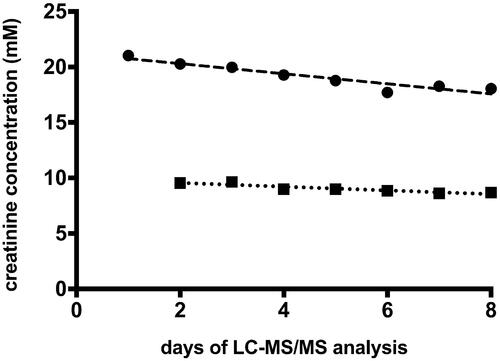
Table 9. Freeze-thaw stability.
Table 10. Descriptive statistics for creatinine and UA measured using reference and index assays.
Figure 6. Passing–Bablok regression analysis of the two methods for UA (A), and creatinine (B). Index test: LC-MS/MS method; reference test: enzymatic colorimetric assay (A), and enzymatic assay (B); diamonds: UA samples (n = 46); squares: creatinine samples (n = 51); green line: regression line; red lines: 95%CI, black line: y = x. Spearman's correlation coefficients (A) 0.964, (p = 5.6582E-27), intercept median (A) −3.539 (95% CI: −28.560 to 24.247), slope (A) 1.057 (95% CI: 1.041–1.074). Spearman's correlation coefficients (B) 0.869 (p = 1.289E-16), intercept median (B) −0.620 (95% CI: −0.979 to −0.161), slope (B) 0.921 (95% CI: 0.886–0.964).
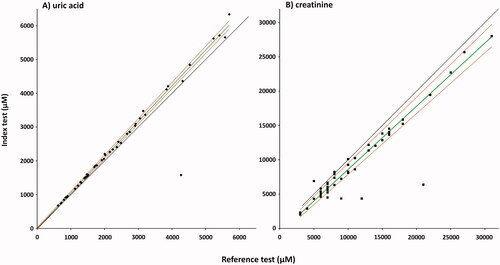
Figure 7. Bland–Altman plot for urinary UA (A), and creatinine (B). Index test: LC-MS/MS method; reference test: enzymatic colorimetric assay (A), and enzymatic assay (B); diamonds: UA samples (n = 46); squares: creatinine samples (n = 51); green line: average delta %; red lines: 1.96SD. The mean bias A: −3.5% (1.96 SD, −32.0 to 25.0%) for the enzymatic colorimetric assay minus LC-MS/MS method; B: 18.6% (1.96 SD, −24.43 to 61.63%) for the enzymatic assay minus LC-MS/MS method.
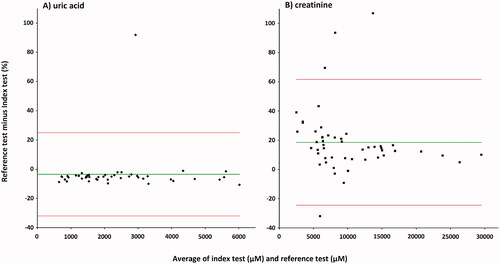
Table 11. Comparison of variation coefficients for UA and creatinine.
Data availability statement
The data that support the findings of this study are available from University Hospital of North Norway (UNN) – but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are, however, available from the authors upon reasonable request and with permission of UNN and the Regional committee for medical and health research ethics.