Figures & data
Table 1. Samples of earthworm guts used for this study
Figure 1. Typical denaturing gradient gel electrophoresis profiles for polymerase chain reaction-amplified fragments of 16S rRNA genes for gut samples from Pheretima hilgendorfi and P. heteropoda collected on 2007 (June or November) and 2008 (June): f, fore-gut; m, mid-gut; h, hind-gut. Numbers with f, m, or h on each lane correspond to individual number. Markers were electrophoresed in Lanes 7, 22, and 23. Bands typically found in most lanes (A–C) are illustrated in the box.
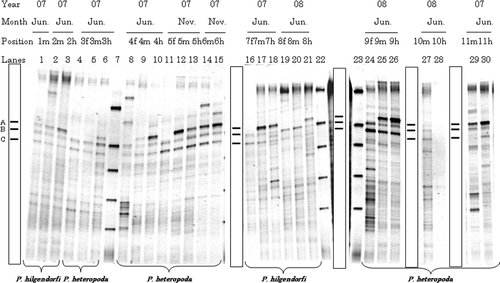
Figure 2. Typical denaturing gradient gel electrophoresis profiles for polymerase chain reaction-amplified fragments of 16S rRNA genes for gut samples from Allolobophora japonica. Explanations for each lane shown in the figure are the same as those in . Markers were electrophoresed in Lanes 31 and 49. Bands typically found in most lanes (a–j) are illustrated in the box.
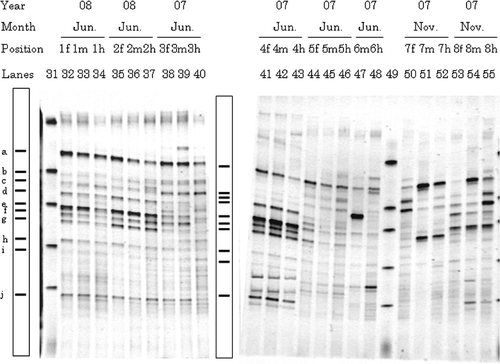
Figure 3. Typical denaturing gradient gel electrophoresis profiles for polymerase chain reaction-amplified fragments of 16S rRNA genes from representative surrounding soils at 2007 and 2008: 0, 0–5 cm depth; 5, 5–10 cm depth; 10, 10–20 cm depth. Numbers in parenthesis behind 0, 5, or 10 correspond to sampling site. Markers were electrophoresed in Lanes 62 and 72.
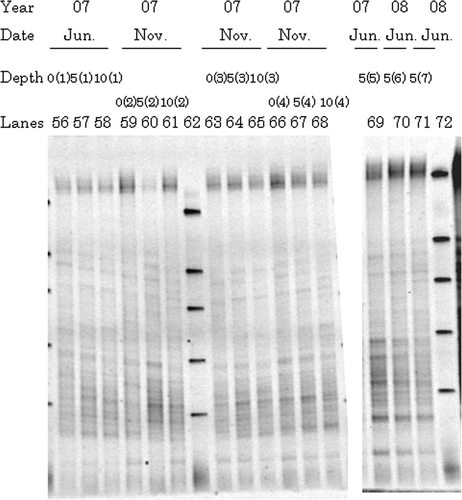
Figure 4. Two-dimensional plot of multidimensional scaling stimulus scores for denaturing gradient gel electrophoresis banding patterns from mid-guts and surrounding soils. Open circle, Pheretima hilgendorfi gut; closed circle, P. heteropoda gut; open triangle, Allolobophora japonica gut; open square, surrounding soil. Numbers correspond to those of the lanes shown in Figs .
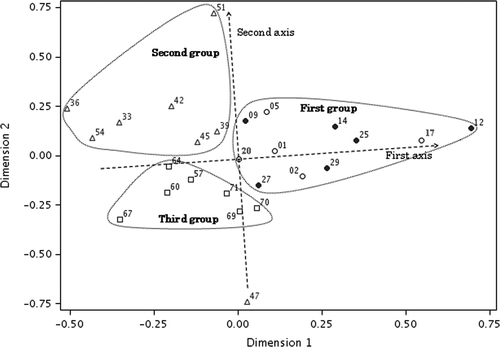
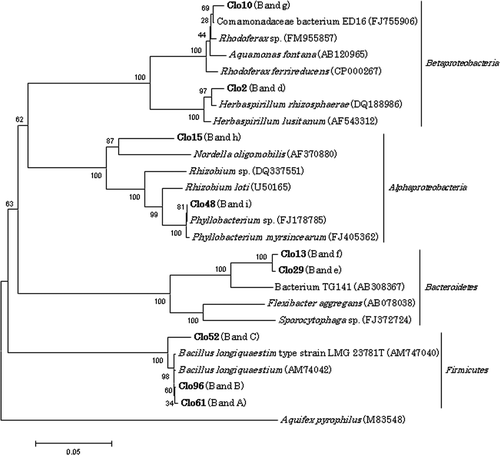
Figure 5. Phylogenetic relationships of 16S rRNA gene sequences for clones which showed the same migration as dominant bands in direct polymerase chain reaction-denaturing gradient gel electrophoresis. Sequences of clones obtained in the present study are shown in boldface. Bootstrap values based on 1000 replications are shown as a percentage at branching points. Bar indicates 0.05 substitutions per nucleotide position.