Figures & data
Table 1. Cycle conditions for the polymerase chain reaction (PCR) amplification with the cpcB-cpcA IGS region and g20 genes
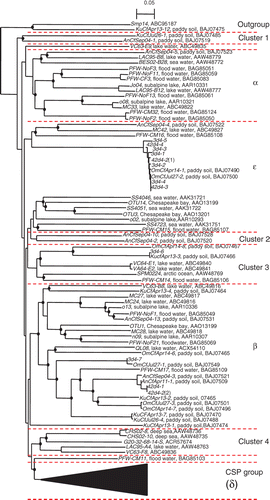
Figure 1. Distribution of polymerase chain reaction (PCR)-product of cpcB-cpcA IGS region genes in different buoyant density fractions. Values in the figure indicate the fraction numbers. M indicates 100-base pair (bp) DNA ladder used as size marker (New England Biolabs). Upper lows, carbon-12 (12C)-callus treatment; Lower lows, carbon-13 (13C)-callus treatment.
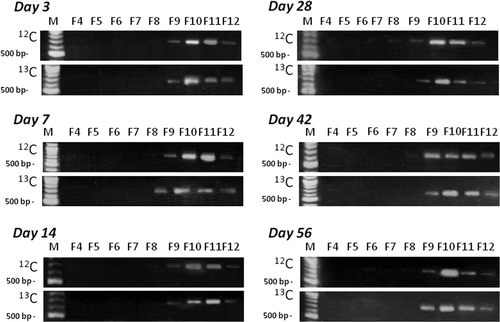
Figure 2. Distribution of polymerase chain reaction (PCR)-product of g20 genes in different buoyant density fractions before cutting. Values in the figure indicate the fraction numbers. M indicates 100-base pair (bp) DNA ladder used as size marker (New England Biolabs). Upper lows, carbon-12 (12C)-callus treatment; Lower lows, carbon-13 (13C)-callus treatment. Arrows represent PCR products about 600-bp in length.
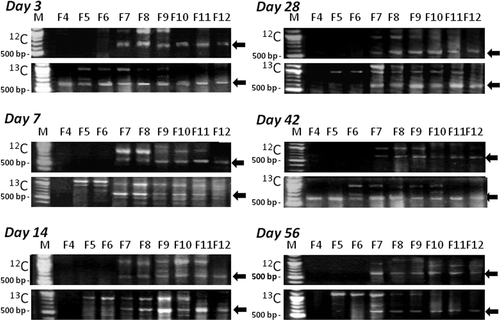
Figure 3. DGGE band patterns of polymerase chain reaction (PCR)-amplified g20 gene in isopycnically fractioned samples (carbon-12 (12C-)- and carbon-13 (13C)-callus treatment). N; non-callus treatment before fractionation. 12 and 13: 12C- and 13C-callus treatment. F4-F12; fraction names, C; unfractionated DNA from samples with 12C-callus added. DGGE band names were assigned as xdy-z in which x, y and z indicate the date of sampling, fraction number and band number, respectively (e.g., 3d4-1 was the band Day 3, the fraction F4 and the first band from the top).
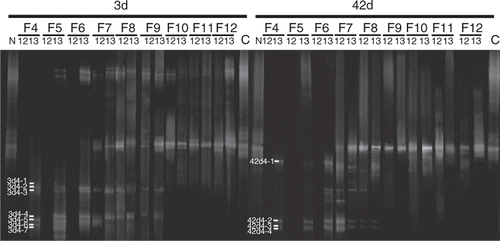
Figure 4. Neighbor-joining phylogenetic tree showing the relationship of g20 amino acid sequences from the paddy field soil in this study. The numbers in the parentheses behind the clone name denote the clone number isolated from same DGGE bands. The closed circle indicates internal nodes with at least 50% bootstrap support.