Figures & data
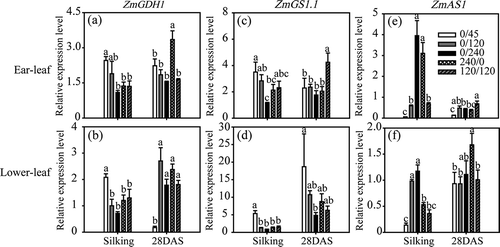
Figure 1 Grain yield, (a) nitrogen (N) accumulation, (b) N utilization efficiency (NUtE), (c) and N harvest index (d) of maize plants (Zea mays L.) grown under different N treatments. Bars indicate means ± standard error (SE) (n = 6). Significant differences at P < 0.05 are indicated by different letters.
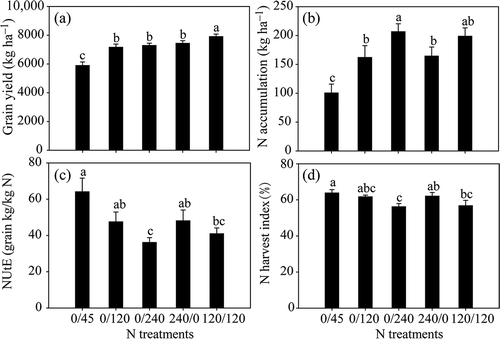
Figure 2 Expression of genes ZmNR1 (a, b), ZmFd-GOGAT1 (c, d) and ZmAspAT1.3 (e, f) in ear leaves (a, c, e) and lower leaves (b, d, f) of maize plants (Zea mays L.) grown under different nitrogen (N) treatments. Transcript levels of ZmNR1, ZmFd-GOGAT1 and ZmAspAT1.3 were normalized to those of the control gene Alpha tubulin4, which were quantified by quantitative real-time polymerase chain reaction (PCR) using gene-specific primers. Bars indicate means ± standard deviation (SD) (n = 3). Significant differences within the same growth stage (silking or 28 days post-silking, DAS) at P < 0.05 are indicated by different letters.
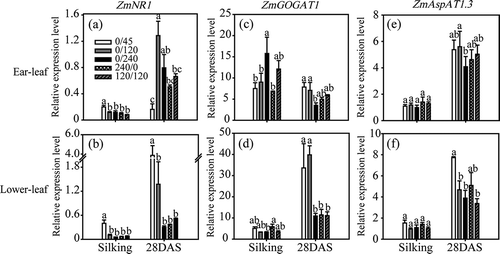
Figure 3 Expression of genes ZmGDH1 (a, b), ZmGS1.1 (c, d) and ZmAS1 (e, f) in ear leaves (a, c, e) and lower leaves (b, d, f) of maize plants (Zea mays L.) grown under different nitrogen (N) treatments. Transcript levels of ZmGDH1, ZmGS1.1 and ZmAS1 were normalized to those of the control gene Alpha tubulin4, which were quantified by quantitative real-time polymerase chain reaction (PCR) using gene-specific primers. Bars indicate means ± standard deviation (SD) (n = 3). Significant differences within the same growth stage (silking or 28 days post-silking, DAS) at P < 0.05 are indicated by different letters.