Figures & data
Table 1. Aboveground biomass, phosphorus (P) concentration, P uptake, and AMF colonization of soybean at the flowering stage†.
Table 2. Soybean grain yield and yield components at the harvest stage†.
Figure 1. DGGE profiles of AMF 18S rRNA genes amplified from the soil and plant roots. Bands indicated with arrows were sequenced, and those with the same alphabet had an identical sequence. The subscript numbers of each band are omitted in other figures. M, DGGE marker IV (Nippon Gene, Tokyo, Japan); CONT, control; NTWC, no-till cultivation after winter wheat cover cropping.
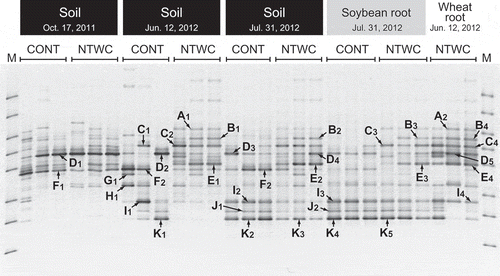
Figure 2. Shannon diversity indices based on the DGGE profiles of soil and root samples. Error bars represent standard errors of triplicate plots. Statistically significant differences were determined using Welch’s t-test and are indicated with **P < 0.01. ns, not significant; CONT, control; NTWC, no-till cultivation after winter wheat cover cropping.
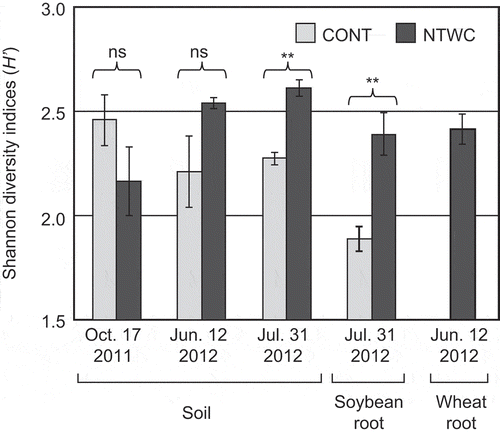
Figure 3. Cluster analysis based on the DGGE profiles of soil and root samples. The Bray–Curtis dissimilarity coefficient calculated from band presence/absence data and the group average method were employed to construct the dendrogram. The rows and columns of heat map represent samples and DGGE bands, respectively. Names of the sequenced bands are indicated on the columns with the closest related family. CONT, control; NTWC, no-till cultivation after winter wheat cover cropping.
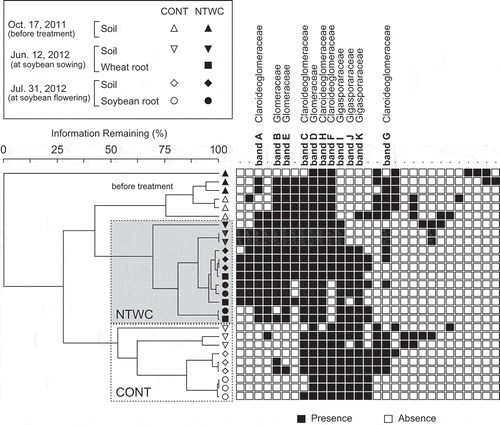
Figure 4. Non-metric multidimensional scaling (NMDS) ordination of AMF communities in soil and soybean root samples collected at the flowering stage (31 July 2012). The NMDS was based on the Bray–Curtis dissimilarity matrix calculated from band presence/absence data of the DGGE profiles. The stress value represents the goodness of fit for the ordination in the reduced dimension (Kruskal Citation1964). CONT, control; NTWC, no-till cultivation after winter wheat cover cropping.
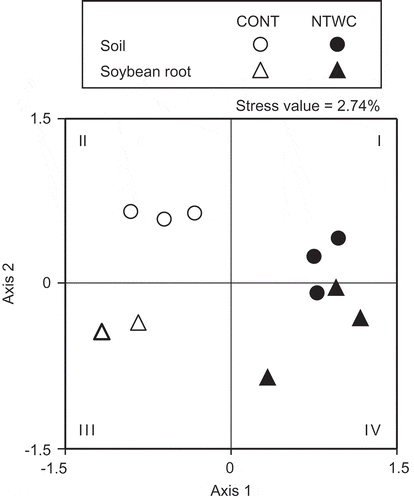
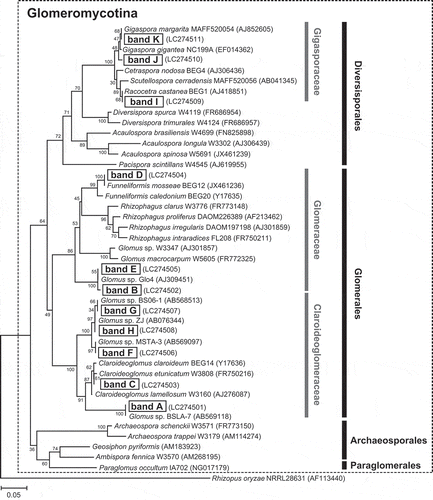
Figure 5. Neighbor-joining tree based on the partial 18S rRNA gene sequences (approximately 240 bp) obtained from the major DGGE bands and from public DNA databases. Names of the DGGE bands are indicated by letters without the subscript number. Numbers on nodes indicate the bootstrap values of 100 trials. The accession number of each sequence is indicated in parentheses. The tree is rooted with the sequence from Rhizopus oryzae (AF113440) as an outgroup.