Figures & data
Figure 1. The molecular structure of gallica acid, ellagic acid, tannic acid, and castalagin. Tannic acid is a gallotannin, a tannin that has a central carbohydrate partly or fully esterified by gallic acid, whereas castalagin is an ellagitannin with ellagic acid ester bonded to the central carbohydrate.
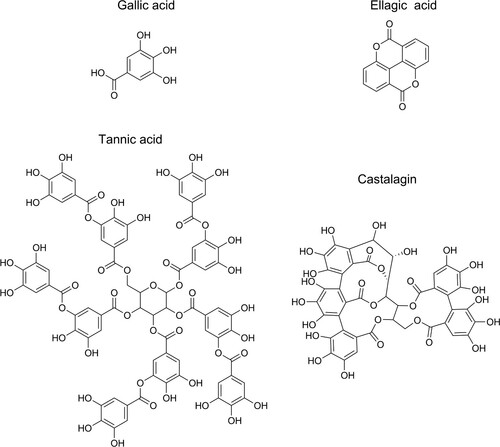
Figure 2. Mechanisms of polyphenolic reactions with iron (with pyrogallol as model compound) adapted from Perron and Brumaghim (Citation2009). Autooxidation (A) by complex formation and subsequent electron transfer in the presence of oxygen leading to a Fe(III)-polyphenol complex. Pro-oxidant reaction (B) by complex formation, iron reduction and semiquinone formation followed by a second iron reduction and formation of a quinone species.
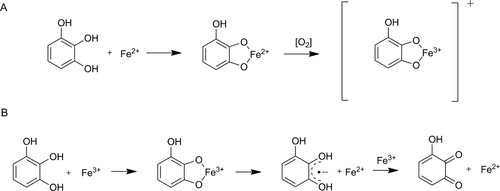
Figure 3. Spatially offset normalised Raman spectra of iron-tannins precipitates synthesised from tannic acid (Fe-TA), extract from archaeological oak (Fe-AOE), and extract from recent oak (Fe-ROE) and of iron-tannin precipitates in The Sword. The band at 1594 cm−1 in the spectrum for The Sword is likely lignin.
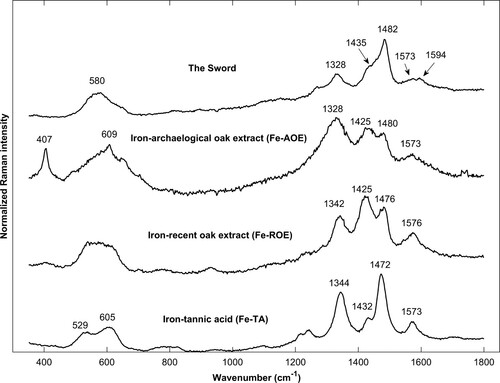
Table 1. Raman band positions (cm−1) for iron-tannin compounds.
Figure 4. Untreated Raman spectra of iron-tannin precipitates identified in different structures in The Sword: filled vessel (top), ray (middle), and cell wall (bottom) in cross section. Signature bands (580, 1328, 1482, and 1573 cm−1) are marked. See micrographs (Supplementary Figures 2–4) showing site of analysis. The top and bottom spectra represent extremes, whereas the middle was most commonly acquired. All spectra exhibit photoluminescence, likely from lignin and degraded wood material.
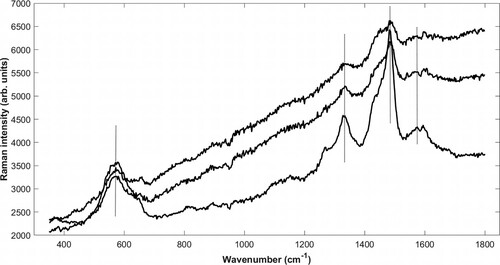
Figure 5. SEM backscattered electron image (A) of a longitudinal section from The Sword showing fibres with partly fractured cell walls that opens up the fibres. EDX mapping of (B) iron and (C) sulphur at the same site. The cell wall has a high iron content but relatively little sulphur.
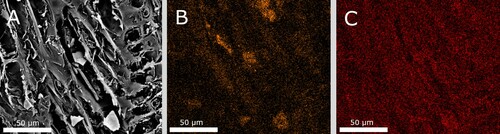
Figure 6. (A) micrograph (100x) of transverse section from The Sword. The wood tissue has turned dark and blackish. Both rays (red arrow), cell walls, and tyloses appears contaminated. In the lower left corner, a small vessel is filled with extensive amount of contaminated material (blue arrow); (B) micrograph (200x) of transverse section from The Sword, showing dark filled lumens of small vessels; and (C), micrograph (630x) of transverse section from The Sword, showing a dark filled vessel and dark contaminated rays.
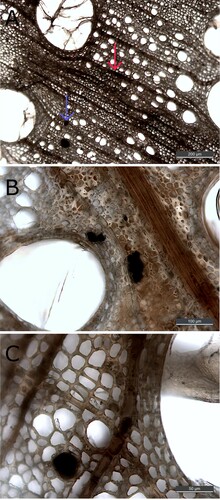
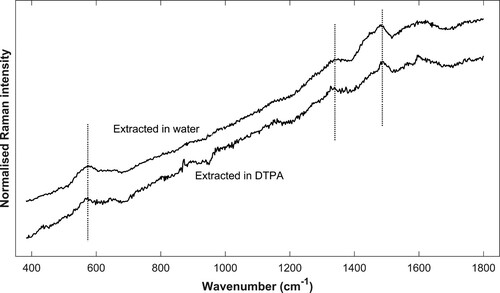
Figure 7. Micrograph (200x) of one cross section from The Sword before (left) and after extraction (right). It is evident that the extraction has led to the removal of depositions and to a lighter appearance; however, Raman analysis still detects iron-tannin precipitates in the remaining dark areas.

Figure 8. Spatially offset normalised Raman spectra. Example spectra of sample extracted in water (top) and DTPA (bottom). The site of analysis is a contaminated ray and a cell wall respectively. Signature bands of iron-tannin (580, 1328, and 1482 cm−1) are marked. As previously described, the band at 1573 cm−1 is often obscured by the presence of lignin, as can be seen in the top spectrum.