Figures & data
Table 1 Computation times (in seconds) of various correlation matrices as a function of the dimension d, for n = 1000 observations.
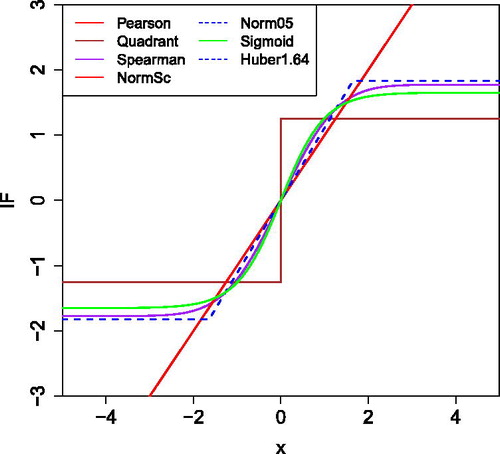
Table 2 Correlation measures based on transformations g with their breakdown value , efficiency, gross-error sensitivity
, rejection point
, and correlation between X and g(X).
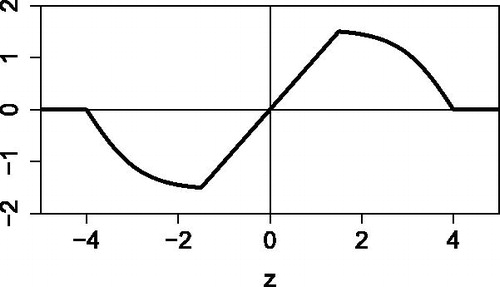
Fig. 3 Illustration of wrapping a standardized sample . Values in the interval
are left unchanged, whereas values outside
are zeroed. The intermediate values are “folded” inward so they still play a role.
![Fig. 3 Illustration of wrapping a standardized sample {z1,…,zn}. Values in the interval [−b,b] are left unchanged, whereas values outside [−c,c] are zeroed. The intermediate values are “folded” inward so they still play a role.](/cms/asset/4af510b5-8ae3-4888-9049-e0e672388910/utch_a_1677270_f0003_b.jpg)
Fig. 4 Bias and MSE of correlation measures based on transformation, for uncontaminated Gaussian data with sample size 100.
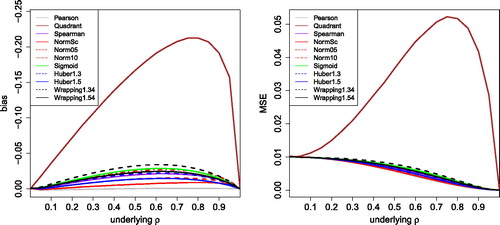
Fig. 5 MSE of the correlation measures in with 10% of outliers placed at k = 3 (left) and k = 5 (right).
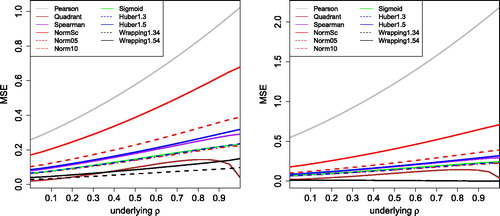
Fig. 6 Bias and MSE of other robust correlation measures, for uncontaminated Gaussian data with sample size 100.
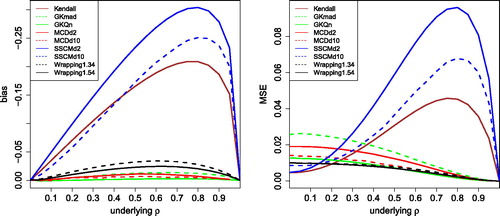
Fig. 7 MSE of the correlation measures in with 10% of outliers placed at k = 3 (left) and k = 5 (right).
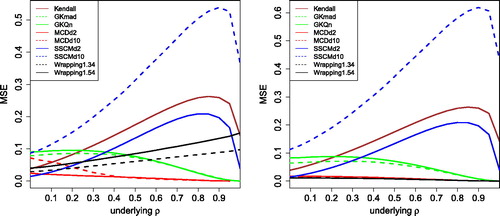
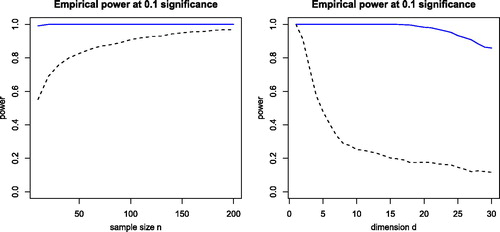
Fig. 8 Left panel: power of dCor (dashed black curve) and its robust version (blue curve) for bivariate X and Y with distribution t(1) and independence except for versus the sample size n. Right panel: power of dCor and its robust version for d-dimensional X and Y with distribution t(1) and n = 100, as a function of the dimension d.