Figures & data
Table 1. Sample types from different mammals sourced between 2007 and 2017 in New Zealand, tested with lipL32, glmU and gyrB PCR to identify pathogenic Leptospira spp.
Figure 1. Flowchart showing selection of DNA samples typed as part of a study of Leptospira spp. infection in mammals in New Zealand. Selection criteria for inclusion in the study were: criteria 1 – samples that were positive to lipL32 PCR; criteria 2 – samples negative to lipL32 PCR but positive to PCR for Leptospira spp. in previous studies; and criteria 3 – samples negative to lipL32 PCR but that came from animals seropositive to Tarassovi in previous studies. Samples were subjected to typing with the speciating PCR (glmU and/or gyrB) if they were positive to any one of these selection criteria.
Figure 2. Leptospira species and serovars identified in DNA samples from New Zealand mammals stratified (A) by the year of sample acquisition and whether they were found in farmed animals or wildlife; and (B) by the mammalian hosts in which they were identified (numbers within each bar show the number of samples from each host in which each Leptospira strain/serovar was identified).
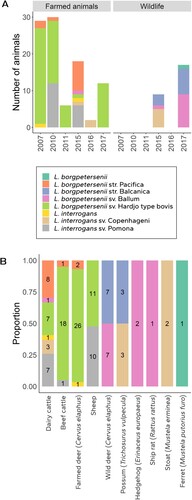
Table 2. An updated summary of Leptospira spp. detected by culture or molecular typing in New Zealand mammals.
