Figures & data
Table 1. Crystal data, data collection, and structure refinement parameters of [Ni(mdeaH2)2](PhCO2)2.
Figure 1. Molecular structure of [Ni(mdeaH2)2](PhCO2)2. Color code: black, red, blue, turquoise, and white spheres represent C, O, N, Ni, and H, respectively.
2. Color code: black, red, blue, turquoise, and white spheres represent C, O, N, Ni, and H, respectively.](/cms/asset/706be66f-641b-4bd4-9323-8b9b2095406d/gcoo_a_2318779_f0001_c.jpg)
Table 2. Selected bond lengths and angles of 1.
Figure 4. Molecular Hirshfeld surfaces (a) dnorm, (b) shape index, (c) curvedness, (d) di and (e) de of 1.
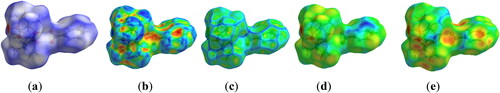
Figure 5. Fingerprint plots of 1 showing the percentages of contact contributions to the total Hirshfeld surface area (a) all, (b) resolved into H–H/H–H interactions, (c) C–H interactions, (d) O–H and (e) C–C type of interactions.
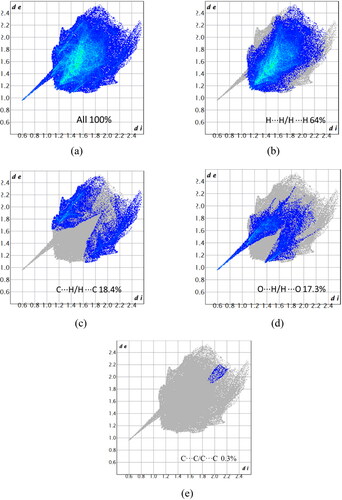
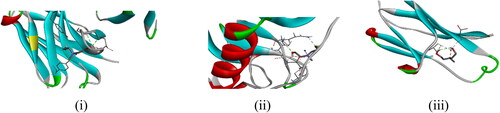
Table 3. The binding energies, inhibition constant, amino acid residues, and bond distances linked with 1.

2 (1).](/cms/asset/3bd4a58d-9708-487a-aa63-f81e4bf5e4ab/gcoo_a_2318779_sch0001_c.jpg)